STED Microscopy Unveiled: A Complete Guide to Imaging Microtubule-Cortex Interactions with Nanoscale Precision
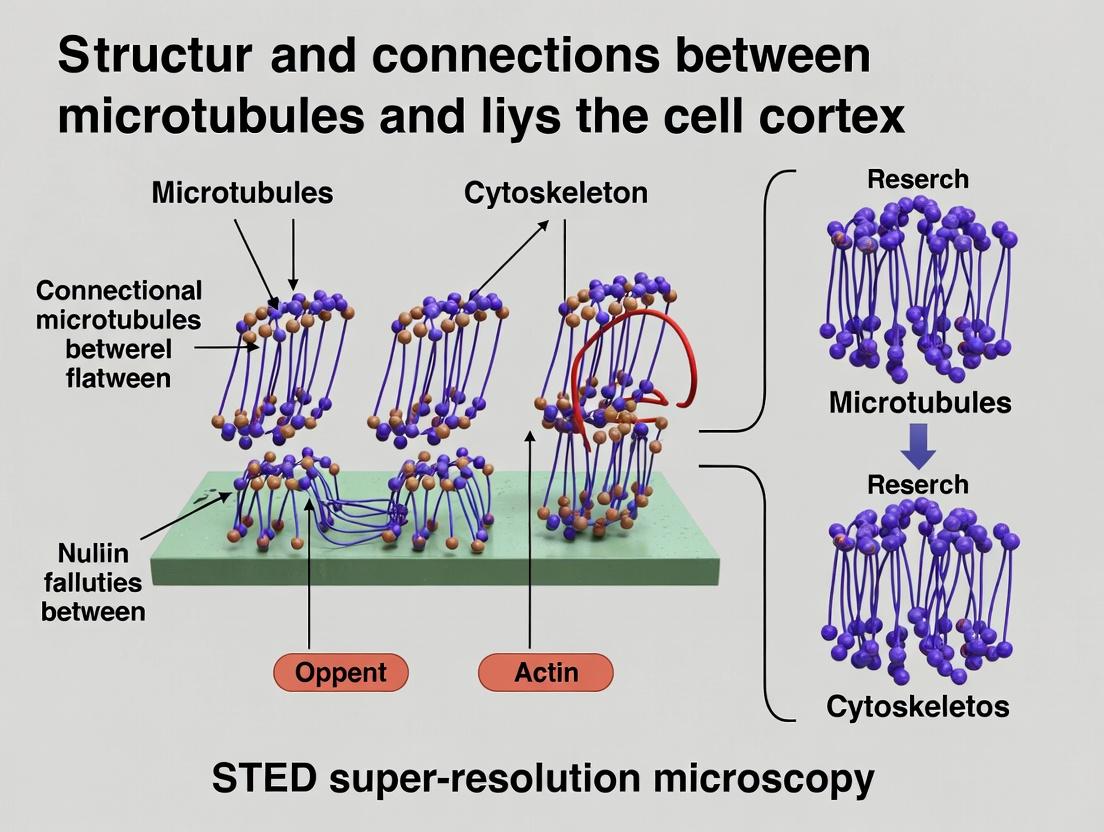
This article provides a comprehensive guide for researchers, scientists, and drug development professionals on applying Stimulated Emission Depletion (STED) super-resolution microscopy to study the nanoscale architecture and dynamics of microtubule-cortex...
STED Microscopy Unveiled: A Complete Guide to Imaging Microtubule-Cortex Interactions with Nanoscale Precision
Abstract
This article provides a comprehensive guide for researchers, scientists, and drug development professionals on applying Stimulated Emission Depletion (STED) super-resolution microscopy to study the nanoscale architecture and dynamics of microtubule-cortex connections. We begin by establishing the biological and structural foundation of these critical cellular junctions. We then detail a complete methodological pipeline for STED sample preparation, imaging, and data acquisition specific to microtubule-cortex interfaces. A dedicated troubleshooting section addresses common challenges in labeling, resolution, and photostability. Finally, we validate STED's performance by comparing it with other super-resolution techniques (e.g., PALM/STORM, SIM) and traditional fluorescence microscopy, assessing its unique advantages and limitations for quantitative analysis in cell biology and cytoskeletal research.
The Nanoscale Nexus: Understanding Microtubule-Cortex Connections and Why STED is Essential
Application Notes
Within the thesis framework of developing and applying STED super-resolution microscopy to study the nanoscale architecture and dynamics of microtubule-cortex connections, these application notes highlight critical biological insights and methodologies. STED microscopy, achieving resolutions below 50 nm, is uniquely positioned to visualize the precise anchorage of microtubule plus-ends to cortical sites and the protein complexes that mediate these interactions, moving beyond the diffraction limit of conventional confocal microscopy.
Key Biological Insights:
- Cell Division (Cytokinesis): Microtubule-cortex connections are essential for the positioning and stability of the actomyosin contractile ring. The centralspindlin complex (MKLP1 kinesin and MgcRacGAP) recruits the RhoA GEF Ect2 to the cell cortex, locally activating RhoA to promote actin contractility. STED imaging has resolved the precise spatial segregation of Ect2 and Anillin within the cortical cytokinetic ring.
- Cell Migration: In polarized migrating cells, microtubules are selectively captured at the cell cortex, particularly at focal adhesions and the leading edge. +TIP proteins (e.g., CLASPs, APC) interact with cortical actin-binding proteins to promote microtubule stabilization, guiding vesicle transport for focal adhesion turnover and membrane protrusion.
- Cell Polarity Establishment: During asymmetric cell division or epithelial polarization, conserved Par complexes (e.g., Par3/Par6/aPKC) are localized to specific cortical domains. These complexes act as cortical landmarks for microtubule capture, ensuring oriented spindle alignment and polarized cargo delivery.
STED-Specific Advantages: The power of STED in this field lies in its ability to:
- Distinguish whether proteins like CLASP2 or NuMA are directly at the microtubule tip, embedded in the cortex, or forming a physical linker.
- Quantify the co-localization efficiency of linker proteins (e.g., Astrin/SPAG5, Kif18b) with both microtubule ends and cortical markers.
- Resolve the nanoscale organization of cortical docking sites during dynamic processes like furrow ingression or leading edge advancement.
Protocols
Protocol 1: STED Imaging of Microtubule Plus-Ends at the Cortex in Migrating Cells
Objective: Visualize the interaction between EB3 (microtubule plus-end marker) and CLASP2 (cortical linker) at the leading edge of a migrating fibroblast.
Materials: See Research Reagent Solutions table.
Method:
- Cell Preparation: Plate serum-starved NIH/3T3 fibroblasts on fibronectin-coated (5 µg/mL) #1.5 high-precision coverslips in a 6-well plate. Allow adhesion for 4 hours.
- Stimulation & Fixation: Stimulate migration with 10% FBS for 90 minutes. Fix cells immediately with pre-warmed (37°C) 4% PFA + 0.1% glutaraldehyde in cytoskeleton buffer (CB: 10 mM MES, 150 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 5 mM glucose, pH 6.1) for 10 minutes.
- Immunostaining:
- Quench with 0.1% NaBH4 in PBS for 7 min.
- Permeabilize with 0.2% Triton X-100 in PBS for 10 min.
- Block with 3% BSA in PBS for 1 hour.
- Incubate with primary antibodies (mouse anti-α-Tubulin, 1:500; rabbit anti-EB3, 1:250; chicken anti-CLASP2, 1:200) in blocking buffer overnight at 4°C.
- Wash 3x with PBS.
- Incubate with secondary antibodies: Goat anti-mouse Abberior STAR 635P (1:200), Goat anti-rabbit Abberior STAR ORANGE (1:200), and Goat anti-chicken ATTO 594 (1:200) for 1 hour at RT. Include 5 µg/mL DAPI if needed.
- Wash 3x and mount in ProLong Glass antifade mountant.
- STED Imaging: Image on a 2-channel STED microscope (e.g., Abberior FACILITY or Leica SP8 STED).
- Use a 100x/1.4 NA oil objective.
- For STAR 635P (Microtubules): Excite at 640 nm, STED depletion at 775 nm.
- For STAR ORANGE (EB3) & ATTO 594 (CLASP2): Excite at 561 nm, STED depletion at 775 nm.
- Acquire sequential z-stacks (30 nm steps) at the cell periphery. Set pixel size to 20 nm.
- Analysis: Use deconvolution software (e.g., Huygens) followed by co-localization analysis (Manders' coefficients) for EB3/CLASP2 within 100 nm of the cell edge, defined by the tubulin signal.
Protocol 2: Live-Cell Imaging of Cortical Microtubule Capture During Mitosis
Objective: Monitor the recruitment of the cortical factor NuMA to the cell cortex during anaphase and its correlation with astral microtubule dynamics.
Materials: See Research Reagent Solutions table.
Method:
- Cell Line Generation: Stably transfect HeLa cells with a BAC vector expressing endogenously tagged NuMA-EGFP. Select with appropriate antibiotic (e.g., G418, 1 mg/mL) for 2 weeks.
- Sample Preparation: Plate NuMA-EGFP HeLa cells in an 8-chamber µ-Slide 48 hours before imaging in FluoroBrite DMEM supplemented with 10% FBS and 25 mM HEPES.
- Microtubule Labeling: 1 hour before imaging, add 100 nM SiR-Tubulin live-cell microtubule probe to the medium.
- Image Acquisition on Confocal (for correlative STED):
- Use a spinning-disk confocal system with environmental control (37°C, 5% CO2).
- Capture dual-channel time-lapse images (EGFP and SiR) every 60 seconds from metaphase through cytokinesis using a 63x/1.4 NA oil objective.
- Identify anaphase cells of interest.
- Correlative STED Fixation & Imaging:
- Immediately at a defined timepoint (e.g., anaphase onset + 5 min), rapidly perfuse the chamber with pre-warmed fixation buffer (PFA/GA as in Protocol 1).
- Process the fixed sample for immunostaining against an additional cortical marker (e.g., anti-GFP to boost NuMA-EGFP signal, and anti-γ-tubulin for centrosomes) as in Protocol 1, Step 3.
- Relocate the same mitotic cell using stage coordinates and acquire 2-color STED images of the cortical region adjacent to the spindle poles to resolve NuMA and microtubule ends.
Visualizations
Cytokinesis Cortical Signaling Pathway
STED Imaging and Analysis Workflow
Research Reagent Solutions
| Reagent / Material | Function / Role in Experiment | Example Product / Identifier |
|---|---|---|
| High-Precision Coverslips (#1.5) | Provides optimal thickness and flatness for super-resolution microscopy, minimizing spherical aberration. | Marienfeld Superior, 0.17 mm thickness. |
| PFA with Glutaraldehyde | Provides rapid fixation while preserving ultrastructure and antigenicity for microtubule and cortex imaging. | Electron microscopy grade, 16% PFA ampules; 25% glutaraldehyde solution. |
| Cytoskeleton Buffer (CB) | A physiological buffer for fixation that preserves microtubule integrity and cortical attachments. | Made in-lab per recipe (see Protocol 1). |
| Primary Antibody: Anti-α-Tubulin | Labels the microtubule polymer for defining the cytoskeletal network. | Clone DM1A (Sigma-Aldrich T9026). |
| Primary Antibody: Anti-EB3 | Marks growing microtubule plus-ends; key for identifying sites of cortical contact. | Clone KT36 (Abcam ab53358). |
| Primary Antibody: Anti-CLASP2 | Labels a key cortical protein that binds microtubule plus-ends and actin. | Polyclonal (Proteintech 20879-1-AP). |
| STED-Optimized Fluorophores | Bright, photostable dyes compatible with STED depletion lasers (e.g., 775 nm). | Abberior STAR 635P, Abberior STAR ORANGE, ATTO 594. |
| ProLong Glass Antifade Mountant | High-refractive index mounting medium that hardens, providing optimal stability for STED imaging. | Thermo Fisher Scientific, P36980. |
| SiR-Tubulin | Live-cell compatible, far-red microtubule probe for correlative live-cell and STED imaging. | Cytoskeleton, Inc., CY-SC002. |
| FluoroBrite DMEM | Low-fluorescence background medium essential for live-cell imaging prior to fixation. | Gibco, A1896701. |
| BAC Transgene: NuMA-EGFP | Provides endogenous-level expression of a crucial cortical mitotic protein for live-cell tracking. | GFP-tagged BAC clone (e.g., from GeneCopoeia). |
Application Notes: STED Microscopy for Cortical Microtubule End-Binding Analysis
Super-resolution STED microscopy is critical for resolving the nanoscale architecture and dynamics of microtubule plus-end interactions with the cortical actin meshwork. These interactions, mediated by specific linker proteins, are central to cellular processes like migration, polarization, and division. The application notes below summarize key quantitative findings enabled by STED.
Table 1: Quantitative Parameters of Key Structural Players at the Cortex
| Protein/Complex | Typical Localization Precision (STED) | Average Distance from MT Plus-End to Cortex (nm) | Binding Lifetime at Cortex (s) | Key Interacting Partners (Cortical) |
|---|---|---|---|---|
| EB1 (+TIP) | ~30-50 nm | 50-150 | 1-3 | APC, CLASPs, Actin (indirect) |
| CLASP2 | ~40-60 nm | 20-100 | 5-15 | LL5β, CLIP-170, Actin |
| Dystonin (BPAG1) | ~50-70 nm | Direct linker (0-20) | >30 (stable) | Plectin, EB proteins, Actin filaments |
| Cytoplasmic Dynein | ~60-80 nm | 0-50 (via dynamic capture) | 2-10 | Dynactin, LIS1, NudE, Cortical NuMA |
| KANK1 | ~40-60 nm | 20-80 | >20 (scaffold) | Talin, Liprin-β, KIF21A |
Table 2: STED Imaging Parameters for Microtubule-Cortex Studies
| Parameter | Recommended Setting | Rationale |
|---|---|---|
| Depletion Wavelength | 775 nm | Optimal for common dyes (e.g., Abberior STAR 635P), minimizes photodamage. |
| Pixel Size | 15-20 nm | Adequate for Nyquist sampling at ~50 nm resolution. |
| Gating | 0.3-1.0 ns | Reduces background fluorescence from scattered light. |
| STED Power | 10-25 mW (at sample) | Balances resolution gain and fluorophore bleaching. |
| Sample Prep | 0.1% Glutaraldehyde + 0.3% PFA | Mild fixation preserves ultrastructure and antigenicity. |
Experimental Protocols
Protocol 1: STED Sample Preparation for Co-visualizing +TIPs and Cortical Actin
Objective: To prepare fixed cells for simultaneous STED imaging of microtubule plus-end binding proteins and the actin cortex. Materials:
- Cells: U2OS or RPE-1 cells grown on high-performance #1.5H coverslips.
- Fixative: 0.3% Paraformaldehyde (PFA) / 0.1% Glutaraldehyde in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9), pre-warmed to 37°C.
- Quenching Solution: 0.1% Sodium Borohydride in PBS for 7 minutes.
- Permeabilization/Blocking Buffer: 0.1% Saponin, 2% BSA, 0.1% Gelatin in PBS.
- Primary Antibodies/Dyes: Mouse anti-EB1 or rabbit anti-CLASP2; Phalloidin conjugated to Abberior STAR 635P; secondary antibodies conjugated to Abberior STAR 580 or STAR RED. Procedure:
- Culture cells on coverslips to 60-70% confluence.
- Rinse briefly in pre-warmed PHEM buffer.
- Fix cells by adding pre-warmed aldehyde fixative for 10 minutes at 37°C.
- Quench autofluorescence with sodium borohydride solution. Rinse 3x with PBS.
- Permeabilize and block with blocking buffer for 1 hour at RT.
- Incubate with primary antibodies diluted in blocking buffer overnight at 4°C.
- Wash 3x for 5 minutes with PBS containing 0.05% Tween-20.
- Incubate with secondary antibodies and phalloidin-635P in blocking buffer for 1 hour at RT in the dark.
- Wash 3x for 5 minutes with PBS.
- Mount coverslips on slides with ProLong Diamond Antifade mountant. Cure for 24 hours before imaging.
Protocol 2: Live-Cell Correlative PALM/STED of Dynein Cortical Recruitmen
Objective: To capture the transient recruitment of cytoplasmic dynein to cortical sites by microtubule plus-ends, using fiduciary markers for STED. Materials:
- Cell Line: HeLa cells stably expressing HaloTag-DYNLL1 (dynein light chain) and EB3-mNeonGreen.
- Dyes: Janelia Fluor 646 HaloTag ligand (for PALM); SiR-Actin (for cortical fiducials).
- Imaging Medium: Leibovitz's L-15 medium without phenol red.
- STED System: Equipped with 775 nm depletion, 592 nm excitation (for JF646), and 640 nm excitation (for SiR). Procedure:
- Seed cells in a glass-bottom dish 24 hours prior.
- Incubate cells with 100 nM JF646 HaloTag ligand and 100 nM SiR-Actin in growth medium for 30 minutes at 37°C.
- Replace with fresh, dye-free medium for 15 minutes to remove unbound dye.
- For live imaging, replace medium with pre-warmed L-15 medium.
- PALM Acquisition: Use TIRF/widefield mode with 640 nm low-power activation and 561 nm readout to track dynein localization over 2-3 minutes.
- STED Acquisition: Immediately after PALM sequence, capture a single high-resolution STED frame of the actin cortex (using SiR, depleted with 775 nm) and a confocal image of EB3 (488 nm excitation). Use the actin STED image as a fiduciary map to correlate PALM localizations.
Diagrams
Title: Protein Pathways Linking Microtubule Plus-Ends to Actin Cortex
Title: STED Sample Prep and Imaging Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Investigating MT-Cortex Linkers
| Reagent | Supplier (Example) | Function in Experiment |
|---|---|---|
| Abberior STAR 635P Phalloidin | Abberior | Direct, high-performance STED-compatible staining of F-actin cortex. |
| HaloTag JF646 Ligand | Promega/Janelia Research Campus | Bright, photostable dye for live-cell PALM/STED tagging of engineered linker proteins (e.g., Dynein subunits). |
| SiR-Actin Kit | Cytoskeleton, Inc. | Live-cell, far-red actin stain for correlative fiduciary marking with minimal perturbation. |
| ProLong Diamond Antifade Mountant | Thermo Fisher Scientific | High-refractive index mounting medium for STED, preserves fluorescence and resolution. |
| PHEM Buffer (100x) | Merck Millipore | Optimized cytoskeleton stabilization buffer for fixation. |
| Mouse anti-EB1 Monoclonal (clone 5/EB1) | BD Biosciences | High-affinity antibody for +TIP staining in fixed-cell STED. |
| Chroma High Performance Filters (e.g., ET775/SP) | Chroma Technology Corp. | Critical for clean depletion beam path in STED system. |
| Fibrillated Nanodiamonds (100 nm) | Adámas Nanotechnologies | Fiducial markers for super-resolution image alignment and drift correction. |
This Application Note details the fundamental resolution limit (~200-250 nm laterally) of conventional fluorescence microscopy and its specific implications for studying the nanoscale interface between microtubules and the actin cortex in cellular mechanobiology. The inability to resolve structures below this diffraction barrier directly impedes research into crucial processes like mitotic spindle positioning, cell polarity, and intracellular transport, which are foundational for understanding cell division and migration in cancer and developmental biology.
Quantitative Comparison of Microscopy Modalities
The following table summarizes the key performance parameters of conventional fluorescence microscopy versus super-resolution techniques, with a focus on requirements for imaging microtubule-cortex connections.
Table 1: Performance Comparison of Microscopy Modalities for Nanoscale Imaging
| Parameter | Conventional Widefield/Confocal | STED Super-Resolution | Implication for Microtubule-Cortex Studies |
|---|---|---|---|
| Lateral Resolution | ~250 nm | ~30-80 nm | Microtubules (~25 nm diameter) are blurred; cortical actin mesh (~50-200 nm pores) is indistinct. |
| Axial Resolution | ~500-700 nm | ~500-600 nm (STED) | Limited Z-discrimination at the thin cortex-membrane interface. |
| Typical Imaging Speed | High (ms-frames) | Moderate (seconds-frames) | Dynamic tracking of plus-end interactions (EB protein comets) at the cortex is challenging with STED. |
| Phototoxicity | Moderate | High (depends on depletion power) | Long-term live-cell imaging of delicate cortical interactions is problematic. |
| Sample/Labeling Demand | Standard fluorophores (e.g., Alexa488) | High-performance dyes (e.g., STAR635) | Requires specific photostable dyes for effective depletion, impacting multicolor labeling strategies. |
| Key Limitation for Cortex Studies | Cannot resolve single microtubule termini contacting cortical actin. | Can resolve single microtubules, but live-cell speed and phototoxicity are concerns. | The nanoscale "connection" (e.g., via dynein, spectraplakins) remains structurally ambiguous without SR. |
Key Experimental Protocol: Assessing Microtubule Proximity to the Cortex Using Conventional Microscopy
This protocol highlights the indirect methods necessitated by the resolution gap.
Protocol Title: Indirect Analysis of Microtubule Cortex-Proximity via Line-Scan Intensity Profiling in Confocal Microscopy.
Objective: To estimate the distance between microtubule plus-ends and the plasma membrane/cortex, despite resolution limits, using fluorescence intensity profiles.
Materials:
- Cells expressing fluorescently tagged microtubule plus-end binding protein (e.g., EB3-GFP) and a cortical/plasma membrane marker (e.g., Lyn-mCherry).
- Confocal microscope with high NA objective (≥1.4) and fast acquisition capabilities.
- Image analysis software (e.g., ImageJ/FIJI, Metamorph).
Procedure:
- Sample Preparation: Plate cells on imaging dishes. Transfert or induce expression of EB3-GFP and Lyn-mCherry. Allow 24-48 hrs for expression.
- Image Acquisition:
- Set up a time-series acquisition with dual-channel simultaneous imaging.
- Set pixel size to ~80 nm/pixel (undersampled relative to diffraction limit) to optimize signal and speed.
- Acquire images at 1-5 second intervals for 2-5 minutes.
- Data Analysis:
- Identify cortical regions with clear microtubule growth trajectories perpendicular to the membrane.
- For each growth event, draw a 5-pixel wide line from the cometing EB3 signal, through the tip, extending into the cortical Lyn signal.
- Generate a kymograph along this line for both channels.
- Plot the fluorescence intensity profile along the line for the frame just prior to microtubule contact/hesitation.
- Measure the distance between the peak of the EB3 signal and the peak/50% intensity of the Lyn signal. This provides an upper-bound estimate of the true gap due to point-spread function (PSF) blurring.
- Statistical Correction: Pool measurements (n>50 events) and apply a deconvolution algorithm (e.g., constrained iterative deconvolution) to estimate the most probable true distance. Note: This correction has inherent uncertainty for gaps <200 nm.
Interpretation & Limitation: Distances measured as <250 nm are unreliable and likely represent direct interactions that cannot be resolved. This method cannot distinguish between a microtubule truly contacting the cortex, terminating 50 nm away, or interacting via a submicroscopic linker protein.
Visualizing the Resolution Challenge and Solution Pathway
Diagram Title: The Resolution Gap from Biological Question to Super-Resolut...
Diagram Title: Comparative Workflow: Confocal vs. STED for MT-Cortex Imaging
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Research Reagent Solutions for Microtubule-Cortex Super-Resolution Studies
| Reagent/Material | Supplier Examples | Function & Critical Role |
|---|---|---|
| STED-optimized Fluorophores | Abberior, Atto, STAR dyes | High photostability and specific excitation/depletion spectra enable effective PSF shrinking. |
| Microtubule Stabilizing Buffer | Cytoskeleton, Inc. | Maintains microtubule integrity during fixation for structural preservation at the cortex. |
| STED-compatible Mounting Medium | ProLong Glass, Abberior Mount | High refractive index, low shrinkage, and anti-fade properties crucial for preserving nanoscale detail. |
| Plasmid: EB3-TagGFP2 | Evrogen, Addgene | A bright, photostable FP variant for live-cell tracking of microtubule plus-end dynamics prior to fixation. |
| Primary Antibody: anti-α-Tubulin, cross-absorbed | Sigma-Aldrich, Abcam | High specificity and affinity for clean microtubule labeling with minimal background at the dense cortex. |
| Phalloidin-ATTO 594 | Sigma-Aldrich, Tocris | High-affinity actin stain for labeling the cortical mesh; ATTO dyes are STED-compatible. |
| Glass-bottom Dishes (#1.5H, 170µm) | MatTek, CellVis | Precise thickness and high optical quality are mandatory for super-resolution microscopy. |
| STED Microscope Alignment Beads | Abberior, TetraSpeck | Fluorescent nanospheres used daily to align and calibrate the excitation and depletion laser beams. |
This application note details the foundational principles and practical protocols for Stimulated Emission Depletion (STED) microscopy, directly supporting the broader thesis research on elucidating the nanoscale architecture and dynamic interactions between microtubules and the actin cortex in cellular mechanics and signaling. Mastering these fundamentals is critical for visualizing sub-diffraction limit structures central to the study.
Core Principles: Breaking the Diffraction Limit
Conventional fluorescence microscopy is limited by diffraction to a resolution of ~200-250 nm laterally. STED microscopy surpasses this limit by employing a physical mechanism to shrink the effective fluorescence volume.
Key Principle: A donut-shaped STED beam (typically at 592 nm, 660 nm, or 775 nm, depending on fluorophore) is co-aligned with the excitation beam (e.g., 488 nm, 561 nm). The STED beam forces excited fluorophores at the periphery of the diffraction-limited spot back to the ground state via stimulated emission before they can fluoresce spontaneously. This confines spontaneous emission to a central sub-diffraction region.
Resolution Equation: The achievable resolution (d) is theoretically unlimited and described by:
d ≈ λ / (2 * NA * √(1 + I/Isat))
where λ is the STED wavelength, NA is the numerical aperture, I is the peak intensity of the STED beam, and Isat is the fluorophore-specific saturation intensity.
Quantitative Performance Data:
Table 1: Typical STED Resolution Performance with Common Fluorophores
| Fluorophore | Excitation λ (nm) | STED λ (nm) | Saturation Intensity (Isat) [MW/cm²] | Achievable Lateral Resolution (with high I) |
|---|---|---|---|---|
| Abberior STAR 488 | 488 | 592 | ~40 | 30-50 nm |
| Abberior STAR 580 | 580 | 660 | ~30 | 40-60 nm |
| Alexa Fluor 594 | 590 | 775 | ~55 | 50-70 nm |
| ATTO 647N | 640 | 775 | ~20 | 30-50 nm |
Table 2: Comparison of Microscopy Modalities for Microtubule-Cortex Imaging
| Modality | Lateral Resolution | Key Advantage for Cortex-MT Research | Key Limitation |
|---|---|---|---|
| Confocal | ~240 nm | Live-cell compatibility, ease of use | Cannot resolve single cortical MTs |
| STED | 30-70 nm | Nanoscale live-cell dynamics; fixed-cell ultra-structure | Higher light dose, specific dyes needed |
| PALM/STORM | 10-30 nm | Highest spatial resolution | Very slow, typically fixed cells only |
Application Notes for Microtubule-Cortex Research
Note 1: Fluorophore Selection: For dual-color imaging of microtubules (labeled with Abberior STAR 580) and cortical markers (e.g., ERM proteins labeled with Abberior STAR 488), use a sequential scan with 660 nm STED for channel 1 and 592 nm STED for channel 2 to minimize crosstalk.
Note 2: Sample Preparation: Optimal fixation for cortical preservation is crucial. Use a brief pre-extraction (0.3% Triton X-100 in PHEM buffer for 30s) followed by 4% PFA + 0.1% Glutaraldehyde fixation for 15 min to maintain cytoskeleton integrity and antigenicity. Quench autofluorescence with 0.1% NaBH₄.
Note 3: Power Optimization: The STED beam power must be carefully titrated. Start at 5-10% of maximum laser power and increase until resolution gain plateaus but before photobleaching accelerates. Typically, 20-40% maximum power (~1-2 mW at sample) provides optimal resolution for live-cell imaging.
Detailed Experimental Protocols
Protocol 1: Immunofluorescence STED Sample Preparation for Fixed Microtubule-Cortex Analysis Objective: To prepare cells for visualizing microtubule ends and cortical actin-associated proteins at super-resolution.
- Culture and Plate: Grow cells (e.g., RPE-1 or HeLa) on high-performance #1.5H coverslips.
- Pre-extraction & Fixation: Aspirate media. Rinse quickly in 37°C PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl₂, pH 6.9). Incubate in PHEM + 0.3% Triton X-100 for 30 seconds. Immediately replace with 4% PFA + 0.1% Glutaraldehyde in PHEM for 15 min at RT.
- Quenching: Rinse 3x in PBS. Incubate in 0.1% NaBH₄ in PBS for 7 min to reduce autofluorescence. Rinse 3x in PBS.
- Blocking & Permeabilization: Block in blocking buffer (3% BSA, 0.1% Triton X-100 in PBS) for 1 hour.
- Primary Antibody Incubation: Incubate with primary antibodies (e.g., mouse anti-α-tubulin, rabbit anti-ezrin) diluted in blocking buffer overnight at 4°C.
- Secondary Antibody Incubation: Rinse 3x in PBS. Incubate with STED-optimized secondary antibodies (e.g., Abberior STAR 580 anti-mouse, Abberior STAR 488 anti-rabbit) at 1:200-1:500 in blocking buffer for 1 hour at RT, in darkness.
- Mounting: Rinse 3x in PBS, then briefly in dH₂O. Mount on glass slide using ProLong Glass antifade mountant. Cure for 48 hours at RT in darkness before imaging.
Protocol 2: Live-Cell STED Imaging of Microtubule Dynamics at the Cortex Objective: To capture the dynamics of microtubule plus-end interactions with the cell cortex.
- Cell Preparation: Transfect cells with a microtubule marker compatible with STED (e.g., SIR-tubulin, or TagRFP-T-EMTB) 24 hours prior.
- Imaging Medium: Use CO₂-independent, phenol-red-free medium supplemented with 10% FBS and 25 mM HEPES.
- System Calibration: Perform daily alignment check using 100 nm crimson fluorescent beads to ensure perfect overlap of excitation and STED donuts.
- Acquisition Settings:
- Use gated detection (time-gating > 0.5 ns) to reduce background.
- Set pixel size to 15-20 nm (1/3 of desired resolution).
- Use pixel dwell time of 5-10 µs.
- For STED beam, apply 660 nm (for SIR/tagRFP) at 20-30% of max power.
- Acquire frames every 2-5 seconds for dynamics.
- Viability Check: Monitor cell health by morphology and cessation of dynamics.
Diagrams
Title: STED Resolution Breaking Workflow
Title: STED Imaging Protocol Flow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for STED Imaging of Microtubule-Cortex Connections
| Item | Function & Rationale | Example Product/Brand |
|---|---|---|
| STED-Optimized Fluorophores | High photostability, high saturation intensity for efficient depletion. | Abberior STAR dyes, ATTO dyes, Chromeo dyes |
| High-Performance Coverslips | #1.5H (170 µm ± 5 µm) thickness for optimal aberration correction. | Marienfeld Superior, Schott Nexterion |
| STED-Compatible Mountant | Low shrinkage, high refractive index (n~1.52), anti-fade properties. | ProLong Glass, Abberior Mount Liquid |
| STED-Tested Secondary Antibodies | Conjugated with optimal dyes, purified for low background. | Abberior STAR SECONDARY, Invitrogen Alexa Fluor STED |
| STED Alignment Beads | 100-200 nm fluorescent beads for daily point spread function (PSF) check and beam alignment. | TetraSpeck microspheres (100 nm), Abberior Calibration Beads |
| Glycerol Objective Correction Collars | For live-cell imaging through plastic dish bottoms, correcting refractive index mismatch. | Built into Leica HC PL APO 100x/1.40 Oil STED White objective |
| Phosphate-Based Extraction/Fixation Buffer (PHEM) | Optimal for cytoskeleton preservation during pre-extraction and fixation. | Prepare in-lab per recipe (60 mM PIPES, 25 mM HEPES, etc.) |
| Live-Cell STED Compatible Dyes | Cell-permeable, bright, photostable dyes for dynamics. | SIR-tubulin, SiR-actin, Janelia Fluor dyes |
Application Notes
STED (Stimulated Emission Depletion) super-resolution microscopy provides a critical advantage for investigating microtubule-cortex connections by uniquely combining direct spatial resolution (not reliant on computational reconstruction) with live-cell compatibility. This enables the quantitative analysis of nanoscale dynamics at the cell periphery where microtubules interact with cortical actin, adhesion complexes, and signaling molecules.
Key Resolved Biological Questions:
- Nanoscale arrangement of microtubule ends (plus-ends) relative to cortical actin networks.
- Dynamics of microtubule contact sites with focal adhesions and adherens junctions.
- Quantification of proteins clustered at microtubule-cortex interaction zones (e.g., +TIP proteins, actin crosslinkers).
- Real-time observation of microtubule growth, shrinkage, and pausing at the cortex under pharmacological perturbation.
Quantitative Performance Data:
Table 1: Comparative Performance of Imaging Modalities for Cortical Microtubules
| Modality | Lateral Resolution (approx.) | Live-Cell Compatibility | Key Limitation for Cortex Studies |
|---|---|---|---|
| Confocal | ~240-280 nm | High | Diffraction limit obscures protein spatial relationships. |
| STED | ~50-80 nm | High | Requires photostable dyes and optimized depletion power. |
| PALM/STORM | ~20-30 nm | Limited (typically fixed) | Slow acquisition; high illumination power unsuitable for prolonged live-cell imaging. |
| SIM | ~100-120 nm | Moderate | Reconstruction artifacts possible at the cortex; lower resolution gain. |
Table 2: Typical STED Imaging Parameters for Live-Cell Microtubule Imaging
| Parameter | Typical Setting/Range | Rationale |
|---|---|---|
| Depletion Wavelength | 592 nm, 660 nm, or 775 nm | Matches dye photophysics (e.g., Abberior STAR 600, SiR-tubulin). |
| Depletion Power | 10-80 mW (at objective back aperture) | Balance between resolution improvement and photodamage. |
| Pixel Size | 15-25 nm | Adequate for Nyquist sampling at ~60 nm resolution. |
| Dwell Time | 2-10 μs | Compromise between signal-to-noise ratio and temporal resolution. |
| Time Interval | 2-30 seconds | Captures microtubule dynamics while minimizing photostress. |
Protocols
Protocol 1: Live-Cell STED Imaging of Cortical Microtubules in Adherent Cells
Objective: To image the nanoscale organization and dynamics of microtubules near the basal cortex.
Research Reagent Solutions & Materials:
Table 3: Essential Materials for Live-Cell STED Imaging of Microtubules
| Item | Function | Example (Product/Supplier) |
|---|---|---|
| Live-Cell Dye | Specific, bright, and photostable labeling of microtubules. | SiR-tubulin (Spirochrome), Abberior LIVE 590 Tubulin (Abberior). |
| Phenol-red free imaging medium | Reduces background fluorescence and autofluorescence. | FluoroBrite DMEM (Gibco), CO₂-independent medium. |
| #1.5 High-Precision Coverslip | Optimal thickness for STED objective lenses (≈0.17 mm). | MatTek dishes or coverslips from Thorlabs, Warner Instruments. |
| STED-compatible mounting system | Maintains cell viability and stability during imaging. | Chamlide magnetic chamber (Live Cell Instrument). |
| STED Microscope | Equipped with 592 nm or 660 nm STED depletion laser and high-sensitivity detectors. | Abberior INSTRUMENTS, Leica Stellaris, or custom setups. |
Procedure:
- Cell Preparation: Seed cells (e.g., U2OS, NIH/3T3) onto a STED-compatible #1.5 glass-bottom dish 24-48 hours before imaging to achieve 50-70% confluence.
- Labeling: Dilute live-cell microtubule dye (e.g., SiR-tubulin) in pre-warmed, phenol-red free medium to the manufacturer's recommended working concentration (typically 100-500 nM). Incubate cells for 1-2 hours at 37°C, 5% CO₂.
- Post-staining Wash: Replace staining medium with fresh, pre-warmed, phenol-red free imaging medium. Incubate for 30 minutes to allow for unbound dye clearance.
- Microscope Setup:
- Mount the dish on the pre-warmed (37°C) stage.
- Use a 100x oil immersion STED objective (NA 1.4).
- Set excitation lines appropriate for the dye (e.g., 640 nm for SiR-tubulin).
- Critical: Align the STED depletion doughnut (e.g., 775 nm for SiR-tubulin) precisely with the excitation focus using a reference sample (e.g., gold nanoparticles).
- Image Acquisition:
- Locate a well-spread cell using low-power confocal mode.
- Select a focal plane at the basal cortex (just above the adhesion plane).
- Set scan parameters: Pixel size 20 nm, dwell time 5 μs.
- Optimization: Start with low STED power (e.g., 10% of max) and increase incrementally until the desired resolution is achieved without visible photodamage (blebbing, microtubule depolymerization).
- For time-lapse, set the interval to 5-10 seconds and limit total acquisition time to 2-5 minutes.
- Data Handling: Save raw data in a non-proprietary format (e.g., .tiff). Process for contrast adjustment and deconvolution (if required) using vendor or open-source software (ImageJ, Fiji).
Protocol 2: Correlative STED and Actin Imaging at the Cortex
Objective: To visualize the spatial relationship between microtubule ends and the actin cortex.
Procedure:
- Follow Protocol 1 steps 1-3 for microtubule labeling.
- Co-labeling Actin: During the final 30-minute wash/incubation, add a live-cell actin stain compatible with STED and spectrally distinct from the microtubule dye (e.g., SiR-actin for 775 nm STED, or Abberior LIVE 510 Actin for 592 nm STED).
- Sequential STED/Confocal Acquisition:
- First channel: Acquire microtubule image in STED mode (e.g., excitation 640 nm, depletion 775 nm).
- Second channel: Immediately switch to acquire actin image. To preserve actin integrity, acquire this channel in confocal mode (no depletion) using the appropriate excitation (e.g., 510 nm).
- Use sequential line scanning to minimize channel crosstalk.
- Registration: Use fluorescent beads or structural features to align the two channels post-acquisition if needed.
Visualizations
Title: Live-Cell STED Imaging Workflow for Microtubules
Title: STED's Role in Microtubule-Cortex Research Thesis
A Step-by-Step STED Protocol for Visualizing Microtubule-Cortex Anchors
This application note provides detailed guidance on fluorophore selection for dual-color STED microscopy, specifically for imaging the interface between microtubules and the actin cortex. Within the broader thesis investigating microtubule-cortex connections in cellular mechanics and signaling, the choice of photostable probes is paramount. Successful imaging requires dyes that withstand high-intensity STED depletion lasers, exhibit minimal cross-talk, and can be efficiently targeted to dynamic protein structures. The following sections synthesize current data and protocols to enable robust experimental design.
Quantitative Comparison of Photostable Dyes for STED
Table 1: Performance Characteristics of Common STED Dyes for Cytoskeletal Labeling
| Fluorophore | Excitation (nm) | STED Depletion (nm) | Emission Peak (nm) | Relative Photostability (τ½) | Recommended Target | Key Advantage |
|---|---|---|---|---|---|---|
| Abberior STAR 635P | 635 | 775 | 650 | Very High (100%) | Microtubules (via antibodies) | Exceptional photostability, ideal for time-lapse STED |
| ATTO 594 | 594 | 775 | 624 | High (~80%) | Cortex (Phalloidin conjugates) | Brightness, good for actin |
| KK114 | 594 | 775 | 670 | Very High (~95%) | Microtubules (SNAP-tag) | High photon yield, low blinking |
| SiR700 | 652 | 775 | 674 | Moderate-High (~70%) | Live-cell microtubules (SiR-tubulin) | Cell-permeant, low toxicity |
| Abberior STAR 580 | 580 | 775 | 605 | High (~85%) | Cortex (Membrane dyes) | Excellent for two-color with 635P |
| ATTO 647N | 645 | 775 | 670 | Moderate (~60%) | General immunolabeling | Widely available, proven |
Note: Relative Photostability is normalized to STAR 635P under identical STED imaging conditions. τ½ refers to the halftime for photobleaching.
Table 2: Dye Pair Selection for Two-Color Microtubule/Cortex STED
| Microtubule Dye | Cortex Dye | Recommended STED Depletion Wavelength | Spectral Crosstalk Risk | Suitability for Live/ Fixed Cell |
|---|---|---|---|---|
| STAR 635P | STAR 580 | 775 nm (for both) | Very Low | Excellent for Fixed |
| KK114 | ATTO 594 | 775 nm (for both) | Low | Fixed |
| SiR700 (live) | SiR-actin (650 ex) | 775 nm & 775 nm | Moderate (requires sequential imaging) | Live-cell only |
| ATTO 647N | ATTO 590 | 775 nm & 775 nm | Low | Fixed |
Detailed Experimental Protocols
Protocol 3.1: Fixed-Cell, Two-Color STED Labeling of Microtubules and Actin Cortex
Objective: To prepare fixed U2OS or HeLa cells for simultaneous super-resolution imaging of microtubules and the subcortical actin network.
Materials (Research Reagent Solutions Toolkit):
| Reagent/Material | Function in Protocol |
|---|---|
| Poly-L-Lysine solution (0.01%) | Coats coverslips for improved cell adhesion. |
| PEM Buffer (100 mM PIPES, 1 mM EGTA, 1 mM MgCl₂, pH 6.9) | Microtubule-stabilizing buffer for fixation and washes. |
| 4% Paraformaldehyde (PFA) in PEM | Fixative that preserves cytoskeleton architecture. |
| 0.1% Glutaraldehyde in PEM | Adds additional crosslinking for superior structural preservation. |
| 0.5% Triton X-100 in PEM | Permeabilization agent for intracellular antibody access. |
| NaBH₄ (1 mg/mL in PBS) | Reduces autofluorescence from glutaraldehyde. |
| Blocking Buffer (5% BSA, 0.1% Triton X-100 in PBS) | Prevents non-specific antibody binding. |
| Primary Antibodies: mouse anti-α-tubulin, rabbit anti-ACTA1 (actin) | Target-specific binding to microtubules and actin. |
| Secondary Antibodies: STAR 635P-conjugated anti-mouse, STAR 580-conjugated anti-rabbit | High-photostability STED dyes for detection. |
| Phalloidin-ATTO 594 (optional) | Alternative direct stain for F-actin. |
| Prolong Diamond Antifade Mountant | Preserves fluorescence and reduces photobleaching. |
| High-Precision Coverslips (#1.5H, 170 µm ± 5 µm) | Essential for optimal aberration-free STED imaging. |
Procedure:
- Cell Seeding: Seed cells on poly-L-lysine coated high-precision coverslips in a 24-well plate. Culture until 60-70% confluent.
- Fixation: Aspirate medium. Rinse quickly with pre-warmed (37°C) PEM buffer. Fix with 4% PFA + 0.1% glutaraldehyde in PEM for 10 minutes at 37°C.
- Quenching & Permeabilization: Rinse 3x with PEM. Incubate with fresh NaBH₄ solution for 7 minutes to reduce aldehydes. Rinse with PBS. Permeabilize with 0.5% Triton X-100 in PEM for 10 minutes at RT.
- Blocking: Incubate with Blocking Buffer for 1 hour at RT.
- Primary Antibody Incubation: Dilute primary antibodies in Blocking Buffer. Apply to coverslip and incubate overnight at 4°C in a humidified chamber.
- Washing: Wash 5x for 5 minutes each with PBS + 0.1% Tween-20 (PBST).
- Secondary Antibody Incubation: Dilute dye-conjugated secondary antibodies (e.g., anti-mouse STAR 635P and anti-rabbit STAR 580) in Blocking Buffer. Incubate for 1 hour at RT in the dark.
- Final Wash and Mounting: Wash 5x for 5 minutes with PBST, followed by a final rinse in distilled water. Blot excess water and mount coverslips on slides using Prolong Diamond. Cure for 24 hours at RT in the dark before imaging.
Protocol 3.2: Live-Cell STED of Microtubules Using SiR-Tubulin
Objective: To perform time-lapse STED imaging of microtubule dynamics in living cells.
Materials:
- Complete cell culture medium without phenol red.
- SiR-tubulin reagent (Cytoskeleton, Inc.).
- Verapamil (optional, to enhance dye loading).
- Leibovitz's L-15 medium without phenol red.
- Live-cell imaging chamber.
Procedure:
- Dye Loading: Prepare a 1 µM working solution of SiR-tubulin in pre-warmed culture medium. Add 10 µM verapamil if needed. Incubate cells for 1-2 hours at 37°C, 5% CO₂.
- Preparation for Imaging: Replace staining medium with fresh, dye-free L-15 medium for imaging. Secure the coverslip in a live-cell chamber maintained at 37°C.
- STED Imaging Settings: Use a 652 nm excitation laser and a 775 nm depletion laser. Keep laser powers minimal to reduce phototoxicity (typical 1-5% STED laser power). Acquire images in time-series mode with intervals ≥ 10 seconds to monitor dynamics.
Visualization Diagrams
Diagram Title: STED Experimental Workflow for Cytoskeleton Imaging
Diagram Title: Logic for Selecting STED Fluorophores
Investigating the nanoscale interface between microtubules and the actin cortex is critical for understanding cell mechanics, division, and migration. Super-resolution microscopy, specifically Stimulated Emission Depletion (STED), enables the visualization of these sub-diffraction limit structures. However, the fidelity of STED imaging is exceptionally dependent on sample preparation. Optimal strategies must balance structural preservation, label density, and fluorophore performance. This document details protocols and reagents for fixed and live-cell STED imaging, tailored for research on microtubule-cortex connections.
Fixation Protocols for Structural Preservation
Proper fixation is paramount for capturing the dynamic architecture of cytoskeletal networks in a static state. The choice of fixative impacts antigenicity, structure, and compatibility with STED dyes.
Paraformaldehyde (PFA) Fixation Protocol (Standard)
- Aim: To crosslink proteins, providing good general structural preservation.
- Materials: Pre-warmed culture medium, 37°C PBS, 4% PFA in PBS (pH 7.4), Quenching Buffer (100 mM Glycine in PBS), Permeabilization/Blocking Buffer (0.1-0.3% Triton X-100, 3% BSA in PBS).
- Method:
- Culture cells on high-precision #1.5H coverslips.
- Aspirate medium and rinse gently with 37°C PBS.
- Fix with 4% PFA for 10-15 minutes at room temperature (RT).
- Quench unreacted PFA with Glycine buffer for 5-10 minutes.
- Rinse 3x with PBS.
- Permeabilize and block for 30-60 minutes at RT.
- Note: Over-fixation with PFA can mask epitopes and increase autofluorescence.
Methanol Fixation Protocol (for Microtubules)
- Aim: To precipitate proteins, often preserving labile microtubule structures and certain epitopes better than PFA.
- Materials: Pre-chilled (-20°C) 100% Methanol, PBS.
- Method:
- Place culture dish on ice. Aspirate medium and rinse briefly with ice-cold PBS.
- Immediately add pre-chilled methanol at -20°C. Incubate for 10 minutes at -20°C.
- Rehydrate and wash cells 3x with PBS for 5 minutes each.
- Proceed to blocking (3% BSA in PBS, permeabilization often not required).
- Caution: Methanol can distort membrane structures.
Quantitative Comparison of Fixation Methods for STED
Table 1: Efficacy of Fixation Methods for Cytoskeletal STED Imaging
| Fixative | Concentration/ Method | Incubation Time | Best For | Pros for STED | Cons for STED |
|---|---|---|---|---|---|
| Paraformaldehyde (PFA) | 4% in PBS | 10-15 min RT | General structure, co-labeling | Good crosslinking, preserves overall architecture | May mask epitopes; can induce shrinkage |
| Methanol | 100%, -20°C | 10 min -20°C | Microtubules, tubulin epitopes | Excellent for tubulin, low background | Destroys membranes, can disrupt cortex |
| PFA + Glutaraldehyde (GA) | 4% PFA + 0.1% GA | 15 min RT | Ultra-structural preservation | Superior crosslinking, finest detail | High autofluorescence; requires quenching (NaBH₄) |
| Glyoxal | 3% in PBS | 30 min RT | Preserving protein conformation | Reduced linkage artifacts, good for antibodies | Less common, requires specific buffer |
Immunostaining Protocols for STED-Grade Resolution
High-density, specific labeling with bright, photostable dyes is non-negotiable for STED.
Direct vs. Indirect Immunostaining Protocol
- Direct: Primary antibody conjugated to STED-compatible dye (e.g., Abberior STAR RED, Alexa Fluor 594).
- Protocol: After fixation/permeabilization, incubate with dye-conjugated primary antibody (1-5 µg/mL in blocking buffer) for 1 hour at RT. Wash 3x 10 min with PBS. Mount.
- Advantage: Lower background, simpler multiplexing.
- Indirect: Primary antibody followed by dye-conjugated secondary antibody.
- Protocol: Incubate with primary antibody (in blocking buffer) for 1 hour at RT or overnight at 4°C. Wash 3x 10 min. Incubate with STED-optimized secondary antibody (e.g., Abberior STAR RED-conjugated, 1:200-1:500) for 45-60 min at RT in the dark. Wash 3x 10 min. Mount.
- Advantage: Signal amplification, wider availability.
Critical Mounting Protocol for STED
- Mounting Medium: Use commercial STED-compatible mounting media (e.g., Abberior Mount Solid, ProLong Glass) or a custom medium (e.g., 97% Glycerol, 2% Mowiol, 1% DABCO).
- Method:
- Apply a small drop (~10 µL) of mounting medium to a clean slide.
- Invert the coverslip with cells and lower it onto the medium, avoiding bubbles.
- Seal edges with clear nail polish or a commercial sealant.
- Cure overnight at RT in the dark before imaging.
- Purpose: Preserves fluorescence, reduces photobleaching, and maintains a stable refractive index.
Live-Cell Compatible Labels for Dynamic Imaging
For observing microtubule-cortex interactions in real time, live-cell compatible probes are essential.
SiR-Tubulin / SiR-Actin Protocol
- Principle: Silicon Rhodamine (SiR)-based probes are cell-permeable, fluorogenic (bright upon binding), and compatible with 650 nm STED depletion.
- Reagents: SiR-tubulin or SiR-actin (Spirochrome/Cytoskeleton Inc.), Verapamil (optional, to enhance uptake).
- Protocol:
- Prepare a 1 µM working solution in culture medium from a 1 mM DMSO stock.
- For enhanced labeling, add 50 µM Verapamil.
- Replace cell culture medium with the staining medium.
- Incubate for 1-2 hours at 37°C, 5% CO₂.
- Replace with fresh, pre-warmed imaging medium (without phenol red) for 30-60 minutes to reduce background.
- Image using a 640 nm excitation laser and a STED depletion laser at ~775 nm.
- Note: Low concentrations (50-250 nM) and minimal light exposure are key to avoiding cytotoxicity.
Expression of Fluorescent Protein (FP) Fusions
- Principle: Transfection or transduction with constructs like Lifeact-GFP (F-actin) or EMTB-3xGFP (microtubules).
- Protocol for STED: For STED, prefer green or red FPs optimized for depletion (e.g., mNeonGreen, mScarlet). Use low-expression systems to avoid artifacts. Image in optimal environmental conditions (37°C, 5% CO₂).
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for STED Sample Preparation in Cytoskeletal Research
| Item | Category | Example Product/Brand | Function in STED Preparation |
|---|---|---|---|
| High-Precision Coverslips | Labware | #1.5H (170 ± 5 µm) | Ensures optimal aberration correction for oil-immersion STED objectives. |
| STED-Optimized Dyes | Fluorophore | Abberior STAR RED, Alexa Fluor 594 | Bright, photostable dyes with well-characterized STED depletion wavelengths. |
| Directly Conjugated Antibodies | Detection | Nanobody-/Fab-dye conjugates | Small size improves labeling density and resolution; reduces linkage error. |
| Live-Cell Probes | Chemical Dye | SiR-tubulin, SiR-actin | Fluorogenic, far-red, low-toxicity probes for dynamic super-resolution imaging. |
| STED Mounting Medium | Imaging Medium | Abberior Mount Solid | Maintains fluorescence brightness and stability under high-intensity STED laser. |
| Cytoskeletal Buffer | Buffer | PEM (100 mM PIPES, 1 mM EGTA, 1 mM MgCl₂) | Stabilizes microtubules during pre-extraction or fixation protocols. |
| Quenching Agents | Chemical | Glycine, Sodium Borohydride (NaBH₄) | Reduces autofluorescence from aldehyde fixatives (especially glutaraldehyde). |
| Environmental Chamber | Hardware | Stage-top incubator (e.g., Okolab) | Maintains live cells at 37°C and 5% CO₂ during extended STED time-lapse imaging. |
Visualized Protocols and Pathways
Title: Fixed vs. Live-Cell STED Sample Preparation Workflow
Title: Molecular Interface of Microtubule-Cortex Connections
Application Notes: Optimizing STED for Microtubule-Cortex Studies
Super-resolution microscopy via STED (Stimulated Emission Depletion) is pivotal for visualizing the nanoscale interface between microtubules and the actin cortex, a critical target in cell mechanics and drug development. Optimal configuration of the depletion laser and detector is essential to resolve sub-diffraction features while preserving sample viability. These notes provide a protocol-centric framework for establishing parameters for live-cell or fixed-cell imaging of microtubule-cortex connections.
1. Core Configuration Principles
The goal is to achieve a resolution of <50 nm to distinguish individual microtubules proximal to the cortical mesh. The depletion laser's effective power and wavelength must be balanced against phototoxicity and fluorophore photophysics. Detector settings must maximize signal-to-noise ratio (SNR) for weak emission signals.
Table 1: Recommended STED Depletion Laser Parameters by Fluorophore
| Fluorophore | Excitation (nm) | Depletion Wavelength (nm) | Depletion Power Range (at sample) | Purpose in Microtubule-Cortex Studies |
|---|---|---|---|---|
| Abberior STAR 635P | 635 | 775 (gated STED) | 10-40 mW | Microtubule labeling (secondary Ab). High photostability for time-series. |
| Alexa Fluor 594 | 590 | 775 | 20-60 mW | Common for beta-tubulin immunofluorescence. Moderate photostability. |
| CF568 | 562 | 775 | 15-50 mW | Bright, photostable. Ideal for co-labeling with green cortical markers. |
| ATTO 647N | 640 | 775 | 10-45 mW | High performance in STED. Suitable for live-cell compatible dyes (e.g., SiR-tubulin). |
| Abberior STAR 580 | 580 | 775 | 15-55 mW | For cortical actin markers (phalloidin conjugates) in dual-color setups. |
Table 2: Detector Settings for Time-Gated Detection (Typical Values)
| Parameter | Recommended Setting | Rationale |
|---|---|---|
| Detector Type | HyD (GaAsP) or APD | High quantum efficiency, low noise. |
| Gating Delay | 0.5 - 1.5 ns | Suppresses early fluorescence photons, improving effective depletion and contrast. |
| Gating Width | 5 - 7 ns | Balances signal collection and background rejection. |
| Pixel Size | 15 - 25 nm | Nyquist sampling for ~50 nm resolution. |
| Dwell Time | 5 - 20 µs | Compromise between image quality and acquisition speed/photo-bleaching. |
| Gain | 100 - 120% (for HyD) | Optimized for weak signal; avoid saturation. |
2. Experimental Protocol: STED Imaging of Microtubule Ends at the Cortex
A. Sample Preparation (Fixed Cells)
- Cell Culture: Plate human RPE-1 or U2OS cells on #1.5 high-precision coverslips.
- Fixation & Permeabilization: Fix with 4% PFA + 0.1% glutaraldehyde in PEM buffer (100 mM PIPES, 1 mM EGTA, 1 mM MgCl₂, pH 6.8) for 10 min at 37°C. Quench with 0.1% NaBH₄. Permeabilize with 0.5% Triton X-100.
- Immunostaining: Incubate with primary antibodies (mouse anti-α-tubulin, rabbit anti-MACF1 or ezrin for cortex anchors) overnight at 4°C. Use secondary antibodies conjugated with STED-optimized fluorophores (e.g., STAR 635P for tubulin, STAR 580 for anchor protein).
B. Instrument Setup & Calibration Protocol
- Alignment: Perform daily STED beam alignment using 40 nm gold beads to ensure co-localization of excitation and depletion foci.
- Depletion Power Calibration:
- Image microtubules labeled with STAR 635P at increasing depletion power (5 mW increments).
- Plot measured filament width (FWHM) vs. depletion power. Select the power where width plateaus (~35-45 nm). Typically, 25-35 mW for fixed samples.
- For live-cell (SiR-tubulin), use the minimum power giving stable super-resolution to reduce photostress.
- Wavelength Selection: Set depletion to 775 nm for all fluorophores in Table 1. For dual-color, verify depletion efficiency for both dyes at this wavelength.
- Detector Optimization:
- Set gating delay to 0.8 ns. Adjust in 0.2 ns steps to maximize depletion effect (loss of signal in donut center).
- Adjust gain so the brightest pixel in a preview scan is at 70-80% of the detector's maximum count to avoid saturation.
- Acquisition: Acquire sequential STED channels. Use a 100x/1.4 NA oil objective. Apply 2x line averaging.
3. The Scientist's Toolkit: Key Reagent Solutions
Table 3: Essential Materials for Microtubule-Cortex STED Imaging
| Item | Function & Specification |
|---|---|
| STED-Optimized Secondary Antibodies (e.g., Abberior STAR, ATTO) | Conjugated to dyes with high depletion cross-section at 775 nm and high photostability. |
| Live-Cell Tubulin Probe (SiR-tubulin / Spy555-tubulin) | Cell-permeable fluorogenic dye for dynamic microtubule imaging in live cells. |
| Mounting Medium (e.g., Abberior Mount Solid Antifade) | Prolongs fluorophore longevity under high-intensity STED light. |
| High-Precision Coverslips (#1.5H, 170 ± 5 µm thickness) | Critical for minimizing spherical aberration, especially with oil objectives. |
| Alignment Sample (40 nm Gold Beads) | For precise co-alignment of excitation and STED laser beams. |
| Fluorescent Bead Sample (100 nm crimson beads) | For daily resolution verification and point spread function (PSF) measurement. |
4. Visualization: STED Configuration and Analysis Workflow
Title: STED Configuration and Imaging Workflow
Title: Parameter Balance in STED Setup
In investigating the dynamic interactions between microtubules and the actin cortex, STED (Stimulated Emission Depletion) super-resolution microscopy is indispensable. It allows visualization of sub-diffraction structural details critical for understanding processes like cell division, migration, and intracellular transport. The core experimental challenge lies in optimizing live-cell imaging parameters to capture rapid biological events without compromising resolution or inducing excessive photobleaching and phototoxicity. This document provides application notes and protocols for achieving this balance.
Core Acquisition Parameters & Quantitative Balancing
The interplay between speed (temporal resolution), spatial resolution, and fluorophore longevity is governed by key hardware and software settings. The following table summarizes the primary parameters, their effects, and recommended starting points for imaging Lifeact (actin cortex) and tagged tubulin (microtubules) in live cells using a gated-STED system.
Table 1: Key Acquisition Parameters for Dynamic Live-Cell STED Imaging
| Parameter | Impact on Speed | Impact on Resolution | Impact on Photobleaching | Recommended Starting Point for Microtubule-Cortex Imaging |
|---|---|---|---|---|
| Pixel Dwell Time | ↓ Longer = Slower scan | ↑ Longer = Better SNR, but potential drift | ↑ Longer = Higher dose, more bleaching | 1.0 - 3.0 µs |
| Pixel Size | ↓ Smaller = More pixels, slower | ↑ Smaller than optical resolution = Oversampling | ↑ More pixels = More exposure, more bleaching | 15-20 nm (STED), 40-60 nm (Confocal reference) |
| Frame Size (px) | ↓ Larger = Slower | Indirect: Larger allows wider FOV | ↑ Larger = More exposure per frame | 1024 x 1024 |
| STED Laser Power | No direct effect | ↑ Higher = Better resolution (saturation) | ↑↑ Dramatically higher = Severe bleaching | 5-30% of max (titrate for each fluorophore) |
| Excitation Laser Power | No direct effect | Indirect via SNR | ↑ Higher = Linear increase in bleaching | 0.5-2% of max (minimize while maintaining SNR) |
| Time Gating (Delay/Width) | No direct effect | ↑ Optimal gating improves effective resolution by filtering early photons | ↓ Can reduce bleaching by using only delayed signal | Delay: 0.5-1.5 ns; Width: 3-6 ns |
| Scanning Mode (Bidirectional/Unidirectional) | ↑ Bidirectional = ~2x Faster | ↓ Bidirectional may have artifacts | Similar effect per pixel | Bidirectional for speed, Unidirectional for high fidelity |
| Averaging (Line/Frame) | ↓ More averages = Slower | ↑ Improves SNR, not native resolution | ↑ More exposure = More bleaching | Avoid for dynamics; use line-scan average (2-4x) if needed. |
Table 2: Protocol-Specific Parameter Sets for Different Experimental Aims
| Experimental Aim | Temporal Resolution Goal | Spatial Resolution Goal | Primary Trade-off Strategy | Typical Frame Rate |
|---|---|---|---|---|
| High-Speed Dynamics (e.g., cortical microtubule contact) | High (~1-5 sec/frame) | Moderate (e.g., ~80 nm) | Reduce STED power, increase pixel size, crop FOV. | 0.2 - 1 Hz |
| Ultra-Resolution Snapshots (e.g., structural detail) | Low | Maximum (e.g., ~50 nm) | Increase STED power, use optimal pixel size, average. | N/A (single time point) |
| Long-Term Time-Lapse (e.g., mitotic progression) | Moderate (~10-30 sec/frame) | Moderate-High | Minimize both excitation and STED power, use gating. | 0.03 - 0.1 Hz |
Detailed Experimental Protocols
Protocol 1: Calibration and Setup for Dual-Color Live-Cell STED
Objective: Establish imaging conditions for simultaneous actin cortex (e.g., Lifeact-SCARLET) and microtubule (e.g., SIR-tubulin) visualization. Materials: See "Scientist's Toolkit" below. Steps:
- Sample Preparation: Seed cells on high-performance #1.5H coverslips. Transfer to imaging chamber. For dual-color, incubate with 100 nM SIR-tubulin and transfection reagent for 2-4 hrs pre-imaging, or transfect with Lifeact-fluorescent protein 24 hrs prior.
- System Alignment: Perform daily alignment of STED donut using 40 nm gold beads or dedicated alignment slides. Verify colocalization of excitation and STED beams.
- Spectral Setup: Define detection windows: 500-540 nm for Abberior STAR ORANGE (mimics SCARLET), 580-630 nm for SIR-tubulin. Configure sequential acquisition to avoid cross-talk.
- Parameter Initialization: Load a confocal configuration (STED laser off). Set pixel size to 60 nm, dwell time 2 µs. Find cells using minimal 488 nm/561 nm excitation power (<1%).
- STED Power Titration: On a region of interest, activate 775 nm STED laser for the red channel (SIR-tubulin). Increment power from 0% to 30% in 5% steps, acquiring a single image. Determine the minimum power yielding desired resolution (FWHM of microtubules < 80 nm). Repeat with 595 nm STED laser for green channel if using a orange/red dye pair.
- Gating Optimization: With optimal STED power, adjust time-gating start delay and width to maximize signal-to-background ratio. Start with a 0.5 ns delay and 6 ns width, then fine-tune.
- Final Live-Cell Settings: Apply determined STED powers. Reduce pixel size to 20 nm. Crop frame to region of interest (e.g., 512 x 512) to achieve target frame rate. Set total acquisition duration and interval.
Protocol 2: Photobleaching Mitigation and Health Monitoring
Objective: To establish a baseline for acceptable laser exposure and monitor cell health during time-lapse. Materials: Cell viability dye (e.g., Sytox Green), CO₂-independent medium, environmental chamber. Steps:
- Pre-imaging Health Check: Include 50 nM Sytox Green in imaging medium. Using confocal mode, briefly scan to confirm all nuclei are Sytox-negative.
- Bleaching Rate Test: On a non-essential sample, perform a time-series with intended parameters. Plot mean fluorescence intensity in the ROI over 50 frames. Calculate decay halftime (T₁/₂). Aim for T₁/₂ > 5x the intended imaging duration.
- Adaptive Power Control: If available, enable "constant photon count" or "brightness preservation" modes that adjust laser power dynamically to compensate for bleaching.
- Environmental Control: Maintain temperature at 37°C ± 0.5°C and use phenol-red free, HEPES-buffered medium to stabilize pH without active CO₂ control.
- Post-imaging Validation: Perform a final confocal scan and Sytox Green check to confirm >95% viability. Compare final structure morphology to initial state.
Diagrams
The Scientist's Toolkit
Table 3: Key Research Reagent Solutions for Live-Cell STED of Cytoskeleton
| Item | Function & Rationale | Example Product/Note |
|---|---|---|
| SIR-tubulin / LiveAct-SCARLET | Cell-permeable, bright, photostable fluorophores for labeling microtubules and actin with excellent STED performance. | Spirochrome SIR-tubulin; Lifeact coupled to Abberior STAR ORANGE. |
| #1.5 High-Performance Coverslips | Coverslips with low autofluorescence and precise thickness (170 µm ± 5 µm) for optimal STED donut formation. | Marienfeld #1.5H High Precision or equivalent. |
| Phenol-Red Free, HEPES-Buffered Medium | Maintains pH without CO₂ during imaging, reducing background fluorescence. | Gibco FluoroBrite DMEM with 20 mM HEPES. |
| Environmental Chamber | Maintains live cells at 37°C and humidity to prevent medium evaporation during long acquisitions. | Okolab or Tokai Hit stage-top incubators. |
| STED Alignment Nanoparticles | Fluorescent beads or dedicated slides for daily verification and alignment of the STED depletion donut. | Abberior or NanoImaging 40 nm Gold Beads, OR Aurora STED alignment slide. |
| Antifade Reagents (for fixed samples) | Reduces photobleaching in fixed-cell super-resolution imaging. Not for live cells. | Abberior Mount, ProLong Diamond. |
| Cell Viability Indicator | Dead cell stain to validate health pre- and post-imaging. | Thermo Fisher Sytox Green (nucleic acid stain). |
This application note details a targeted super-resolution microscopy investigation within the broader thesis research on "Nanoscale Mapping of Microtubule-Cortex Anchoring Complexes Using STED Microscopy." The dynamic interaction between microtubule plus-ends and the cell cortex, mediated by proteins like the end-binding protein EB1 and the gamma-tubulin complex component GCP2, is critical for cell polarity, division, and migration. Understanding the precise spatial organization of these proteins at cortical contact sites, below the diffraction limit, is essential for mechanistic models of force transmission and regulatory signaling. This study applies two-color STED nanoscopy to resolve the nanodistribution of EB1 and GCP2 at these strategic locations.
Live-cell and fixed-cell STED imaging of human RPE-1 cells expressing fluorescently tagged EB1 and GCP2 revealed distinct nanoscale patterns at cortical microtubule interaction sites.
Table 1: STED Resolution Performance and Protein Distribution Metrics
| Parameter | EB1 (mNeonGreen) | GCP2 (mScarlet-I) | Measurement Notes |
|---|---|---|---|
| Achieved STED Resolution (FWHM) | 58 ± 3 nm | 62 ± 4 nm | Measured on 40nm gold beads under same conditions. |
| Localization Precision | 12 ± 5 nm | 15 ± 6 nm | Calculated from repetitive measurements of single molecules. |
| Typical Diameter of EB1 Comets | 85 ± 15 nm | N/A | At microtubule plus-ends in the cytoplasm. |
| Cortical EB1 Cluster Diameter | 120 ± 25 nm | N/A | At sites of microtubule-cortex contact. |
| Cortical GCP2 Cluster Diameter | N/A | 95 ± 20 nm | Often observed as discrete puncta. |
| Average EB1-GCP2 Edge-to-Edge Distance | 45 ± 20 nm | At co-occurring cortical sites (n=127 sites). | |
| Frequency of Co-occurrence | 68% of cortical EB1 sites | Percentage of EB1 puncta with a GCP2 puncta within 100 nm. |
Table 2: Experimental Conditions and Reagents
| Component | Specification/Product ID | Purpose/Function in Study |
|---|---|---|
| Cell Line | hTERT RPE-1 | Model human epithelial cell line with stable, non-transformed karyotype. |
| Plasmids | pCMV-EB1-mNeonGreen, pCMV-GCP2-mScarlet-I | For live-cell expression of target proteins. Cloned via Gibson Assembly. |
| Fixative | 4% Formaldehyde + 0.1% Glutaraldehyde in PHEM Buffer | Provides rapid fixation while preserving ultrastructure for STED. |
| Primary Antibody | Mouse anti-α-Tubulin (DM1A) | Microtubule backbone staining for correlation. |
| STED Dye (Secondary) | Goat anti-Mouse IgG, Abberior STAR 635P | High-performance STED-compatible dye for 775nm depletion. |
| Mounting Medium | ProLong Glass Antifade Mountant | High-refractive index, hard-setting medium optimal for STED. |
| STED Microscope | Abberior INSTRUMENTs FACILITY 4color-2D-STED | Equipped with 775nm depletion laser, 595nm & 640nm excitation lines. |
Detailed Experimental Protocols
Protocol: Sample Preparation for Correlative STED Imaging of EB1/GCP2
Objective: Prepare fixed-cell samples expressing fluorescent fusion proteins for two-color STED and immunofluorescence.
- Cell Culture & Transfection: Plate RPE-1 cells on #1.5H high-performance coverslips. At 50-60% confluency, co-transfect with 250 ng each of pCMV-EB1-mNeonGreen and pCMV-GCP2-mScarlet-I using a lipid-based transfection reagent. Incubate for 18-24h.
- Fixation: Rinse cells twice with pre-warmed PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 4 mM MgSO₄, pH 6.9). Fix with 4% formaldehyde + 0.1% glutaraldehyde in PHEM for 10 min at 37°C.
- Quenching & Permeabilization: Rinse 3x with PBS. Quench autofluorescence with 0.1% sodium borohydride in PBS for 7 min. Rinse, then permeabilize with 0.5% Triton X-100 in PBS for 10 min.
- Immunostaining (Microtubules): Block with 5% BSA in PBS for 1h. Incubate with anti-α-Tubulin (DM1A, 1:500 in 1% BSA/PBS) overnight at 4°C. Wash 5x over 1h with PBS. Incubate with Abberior STAR 635P-conjugated secondary antibody (1:500) for 1h at RT in the dark. Wash 5x over 1h.
- Mounting: Rinse coverslips with distilled water and mount on glass slides using ProLong Glass Antifade Mountant. Cure for 48h at RT in the dark before imaging.
Protocol: Two-Color STED Acquisition Parameters
Objective: Acquire super-resolved images of EB1, GCP2, and microtubules with optimal signal-to-noise and resolution.
- System Setup: Use a 100x/1.4 NA oil immersion objective. Align the 775 nm STED depletion donut for both green (mNeonGreen) and red (mScarlet-I) channels using 40 nm fluorescent beads.
- Channel Sequential Acquisition:
- Channel 1 (STED - EB1): Excitation: 595 nm pulsed laser (5 µW at sample). Depletion: 775 nm (max 40 mW at sample). Detection: 610-630 nm bandpass.
- Channel 2 (STED - GCP2): Excitation: 640 nm pulsed laser (8 µW at sample). Depletion: 775 nm (max 45 mW at sample). Detection: 650-720 nm bandpass.
- Channel 3 (Confocal - Microtubules): Excitation: 640 nm (2 µW). Detection: 650-720 nm (STED laser OFF).
- Acquisition Settings: Pixel size: 20 nm. Pixel dwell time: 10 µs. Line accumulation: 4. Use gated detection (delay: 0.5 ns, width: 6 ns) to suppress background fluorescence.
- Control Acquisition: Acquire a confocal image (all STED lasers OFF) of each channel in the same region for comparison.
Protocol: Image Analysis & Nanoscale Colocalization
Objective: Quantify protein cluster dimensions and intermolecular distances.
- Deconvolution & Background Subtraction: Apply a mild deconvolution algorithm (e.g., Richardson-Lucy, 10 iterations) to raw STED data using a measured point spread function (PSF). Subtract constant background based on image mean intensity outside cells.
- Cluster Segmentation: For each channel, apply a bandpass filter and use a Laplacian of Gaussian (LoG) blob detector to identify protein clusters. Set a minimum intensity threshold 3x above local background.
- Distance Measurement: For segmented EB1 and GCP2 clusters within 300 nm (confocal limit), fit a 2D Gaussian to each STED-resolved cluster to determine centroid position. Calculate the edge-to-edge distance as: Centroid Distance - (Radius_EB1 + Radius_GCP2), where radii are derived from the FWHM of the Gaussian fit.
Diagrams & Workflows
Diagram Title: STED Sample Prep and Imaging Workflow
Diagram Title: EB1 and GCP2 at MT-Cortex Interface
The Scientist's Toolkit: Essential Research Reagents & Materials
| Item/Category | Example Product/Identifier | Critical Function in STED Experiment |
|---|---|---|
| Fluorescent Proteins | mNeonGreen, mScarlet-I | Bright, photostable tags ideal for live-cell and fixed-cell STED. |
| STED-Optimized Dyes | Abberior STAR 635P, Atto 594 | Engineered for high saturation intensity and photostability under depletion laser. |
| High-NA Objective | 100x/1.40 NA Oil STED Objective | Essential for collecting maximal signal and achieving theoretical resolution limits. |
| STED Depletion Laser | 775 nm (Pulsed or CW) | Creates the "donut" to deplete periphery of excitation spot, enabling nanoscale resolution. |
| Antifade Mountant | ProLong Glass, Abberior Mount Liquid | Preserves fluorescence over repeated scanning; correct refractive index minimizes aberrations. |
| Image Analysis Software | Fiji/ImageJ with custom macros, ImSpector | For deconvolution, quantitative analysis, and distance measurements of STED data. |
| Cell Culture Substrate | #1.5H High-Precision Coverslips (170µm ±5µm) | Consistent thickness is non-negotiable for optimal STED donut formation and PSF. |
Solving Common STED Imaging Challenges for Cytoskeletal Research
Within a thesis investigating microtubule-cortex connections using STED super-resolution microscopy, managing photodamage is paramount. STED's high-intensity depletion laser exacerbates fluorophore photobleaching and induces cellular phototoxicity, distorting delicate cytoskeletal interactions. This document provides practical application notes and protocols for preparing imaging media and buffers that enhance fluorophore stability and cell viability during demanding super-resolution sessions.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent/Material | Primary Function & Explanation |
|---|---|
| ROXS (Reducing and Oxidizing System) | A chemical cocktail (e.g., Ascorbic acid + Methylviologen) that mitigates photobleaching by quenching triplet states and reactive oxygen species (ROS) before they damage the fluorophore. |
| Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) | A vitamin E analog that acts as a potent antioxidant, scavenging free radicals and significantly reducing photobleaching rates, especially for organic dyes. |
| Cyclooctatetraene (COT) & 1,4-Diazabicyclo[2.2.2]octane (DABCO) | Common anti-fade agents. COT reduces blinking and bleaching by triplet-state quenching. DABCO reduces photobleaching, often used in glycerol-based mounting media. |
| Glucose Oxidase/Catalase (GLOX) System | An oxygen-scavenging system. Glucose oxidase consumes oxygen, limiting the production of singlet oxygen, a key mediator of phototoxicity and bleaching. |
| Cysteamine (MEA) / Cysteamine HCl | A reducing agent used in STORM buffers. It acts as a thiol to maintain dyes in a dark state, but its concentration must be optimized to balance blinking and cell health. |
| Pluronic F-127 | A non-ionic surfactant used to facilitate the delivery of hydrophobic dyes (e.g., certain live-cell probes) into cellular membranes without excessive toxicity. |
| Hanks' Balanced Salt Solution (HBSS) with HEPES | A physiologically balanced salt solution for live-cell imaging. HEPES maintains pH without CO2 buffering, crucial for open-dish microscopy. |
| Phenol Red-Free Medium | Standard cell culture media often contains phenol red, which can autofluoresce and generate ROS under laser illumination, increasing background and photodamage. |
Quantitative Comparison of Anti-fade/Buffering Systems
Table 1: Performance of Common Additives in Live-Cell STED Imaging of Microtubules.
| Additive/System | Typical Concentration | Target Fluorophore(s) | Key Benefit | Reported Increase in Fluorescence Lifetime* | Potential Drawback for Live Cells |
|---|---|---|---|---|---|
| Trolox | 1-2 mM | ATTO 488, STAR 580, Alexa Fluor dyes | Strong radical scavenging | ~2-5 fold | Can affect cell metabolism at high conc. |
| GLOX System | Gluc Ox 0.5 mg/mL, Cat 40 µg/mL, Glucose 10 mM | Most dyes in aqueous env. | Oxygen removal, reduces phototoxicity | ~3-7 fold | Can acidify medium; requires glucose. |
| ROXS (Asc./M.V.) | Ascorbate 1-2 mM, Methylviologen 1 µM - 1 mM | Cy dyes, ATTO dyes | Efficient triplet-state quenching | ~5-10 fold | Methylviologen is toxic; for fixed cells only. |
| Cysteamine (MEA) | 50-150 mM | STORM dyes (e.g., Alexa 647) | Enables blinking for localization | N/A (for blinking) | High osmolality & toxicity; not for long-term live-cell. |
| Commercial Anti-fade Mountants | As per mfr. | All (fixed samples) | Ease of use, often proprietary | Varies widely (~2-10 fold) | Mostly for fixed samples only. |
*Increase in number of detected photons or frames before bleaching under comparable illumination. Values are illustrative from literature.
Experimental Protocols
Protocol 1: Preparing a Live-Cell Imaging Medium for STED of Microtubule-Cortex Probes
Objective: To create an imaging medium that supports cell viability while minimizing photobleaching of a tubulin-labeling dye (e.g., SiR-tubulin) during STED time-lapse. Materials: Phenol red-free imaging medium (e.g., Leibovitz's L-15), HEPES (20 mM, pH 7.4), Trolox (from 100 mM stock in DMSO), Pluronic F-127 (for dye preparation), SiR-tubulin stock. Procedure:
- Prepare a 100 mM Trolox stock solution in DMSO. Store aliquots at -20°C, protected from light.
- On the day of imaging, prepare the working imaging medium. To 50 mL of pre-warmed (37°C), phenol red-free L-15 medium, add HEPES to a final concentration of 20 mM.
- Add Trolox stock solution to a final concentration of 1 mM (e.g., 500 µL of stock to 50 mL). Mix gently.
- Dye Loading: Prepare a 1 µM SiR-tubulin solution in the prepared Trolox medium using the manufacturer's protocol, which may include a brief incubation with 0.1% Pluronic F-127.
- Replace the culture medium on your prepared cells (e.g., COS-7 or HeLa) with the dye-containing/Trolox medium. Incubate for 1-2 hours under standard growth conditions.
- Prior to imaging, replace the loading medium with fresh Trolox-supplemented, dye-free imaging medium to reduce background.
- Proceed with STED imaging, maintaining stage temperature at 37°C.
Protocol 2: Optimizing a GLOX-based Oxygen-Scavenging Buffer for Fixed-Cell STED
Objective: To prepare an anti-fade mounting medium for imaging fixed samples of cortical actin and microtubules (e.g., labeled with Phalloidin-Atto 590 and anti-α-tubulin-Abberior STAR 635) to maximize signal retention. Materials: Glycerol, Tris-Cl (pH 8.0), Glucose Oxidase (from Aspergillus niger), Catalase (from bovine liver), Glucose, β-Mercaptoethanol (optional), #1.5 High-Precision Coverslips. Procedure:
- Prepare a 10x GLOX stock solution:
- 10 mg/mL Glucose Oxidase in 50 mM sodium acetate, pH 5.2. Centrifuge briefly (2 min, 13,000 x g) to pellet insoluble aggregates. Use supernatant.
- 5 mg/mL Catalase in 50 mM Tris-HCl, pH 8.0. Centrifuge as above.
- Aliquot and store at -20°C.
- Prepare a Mounting Buffer Base: 90% (v/v) glycerol, 20 mM Tris-Cl, pH 8.0. Mix thoroughly.
- On the day of mounting, prepare the working GLOX mounting medium:
- To 1 mL of Mounting Buffer Base, add:
- 10 µL of 10x GLOX Glucose Oxidase stock (final ~0.1 mg/mL)
- 10 µL of 10x GLOX Catalase stock (final ~0.05 mg/mL)
- 18 mg of D-(+)-Glucose (final ~100 mM)
- Mix thoroughly but gently by inversion. Avoid introducing bubbles.
- To 1 mL of Mounting Buffer Base, add:
- (Optional) For additional reducing power, add β-Mercaptoethanol to a final concentration of 10-50 mM.
- Apply 10-20 µL of the GLOX mounting medium to a clean slide.
- Invert the coverslip with the fixed and immunolabeled sample onto the medium, avoiding bubbles. Seal the edges with nail polish or a commercial sealant.
- Allow the mountant to set for 10-15 minutes before proceeding with STED imaging. For best results, image within 24-48 hours.
Visualization: Pathways and Workflows
Title: Mechanisms of Photobleaching and Antioxidant Protection
Title: Live vs Fixed Cell STED Preparation Workflows
Optimizing Signal-to-Noise Ratio (SNR) in Dense Cortical Regions
Application Notes & Protocols Thesis Context: Advancing STED super-resolution microscopy to elucidate nanoscale architecture and dynamic interactions between microtubule plus-ends and the actin cortex in cellular processes like division and migration. Optimizing SNR is paramount for reliable, quantitative imaging in these dense, signal-rich regions.
The following parameters critically influence SNR in STED imaging of cortical regions.
Table 1: Primary Factors Affecting SNR in Cortical STED Imaging
| Factor | Optimal Range/Type | Impact on SNR | Rationale |
|---|---|---|---|
| Fluorophore Choice | High photostability, STED-compatible (e.g., Abberior STAR RED, Alexa Fluor 594) | High | Red-emitting dyes reduce background autofluorescence and require lower STED laser power. |
| Labeling Density | Sparse, specific labeling via immunostaining or SNAP/CLIP-tags | High | Reduces intermolecular distance below resolution limit, minimizing apparent density and background. |
| STED Laser Power | 10-80% of saturation power (system-dependent) | Medium-High | Higher power increases depletion but also photobleaching and background. Must be titrated. |
| Excitation Laser Power | Minimal to achieve sufficient signal (1-10 µW at sample) | High | Lower power reduces photobleaching and nonlinear background signals. |
| Pixel Size & Dwell Time | Pixel: 10-20 nm; Dwell: 5-20 µs | Medium | Oversampling improves fidelity but increases acquisition time and photodamage. |
| Sample Preparation | Refractive-index matched mounts (e.g., ProLong Glass) | High | Reduces spherical aberration and scattering, improving signal collection. |
| Gating (Time-Gating) | 0.5-6.0 ns delay post-excitation | Medium | Effectively removes fluorescence from excited-state lifetime of background. |
Table 2: Typical SNR Outcomes with Different Protocols
| Protocol Modifications | Baseline SNR (Confo.) | Achieved SNR (STED) | % Improvement | Key Limitation |
|---|---|---|---|---|
| Standard Immunofluorescence | 8:1 | 5:1 | -37% | Dense labeling, high background. |
| Sparse Labeling + STAR RED | 10:1 | 25:1 | +150% | Requires genetic manipulation or optimized staining. |
| Sparse Labeling + Gated STED | 10:1 | 40:1 | +300% | Increased acquisition complexity and analysis. |
| Recommended: Combined Optimal (Sparse, STAR RED, Gated, RI-matched mount) | 12:1 | 50:1 | +317% | Protocol length and cost. |
Detailed Experimental Protocols
Protocol 2.1: Sparse Labeling of Microtubule Plus-Ends in Fixed Cells
Objective: To achieve low-density, high-specificity labeling of EB3 or CAP-Gly domain proteins marking microtubule plus-ends near the cortex. Reagents: See Toolkit (Section 4). Procedure:
- Culture cells on high-precision #1.5H glass-bottom dishes.
- Fix with pre-warmed 4% PFA + 0.1% glutaraldehyde in cytoskeleton buffer (CB: 10 mM MES, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM MgCl2, pH 6.1) for 10 min at 37°C.
- Quench with 0.1% NaBH4 in PBS for 7 min, twice.
- Permeabilize with 0.2% Triton X-100 in PBS for 10 min.
- Block with 5% BSA, 0.1% Fish Skin Gelatin in PBS for 1 hr.
- Primary Antibody Incubation: Dilute anti-EB1 or anti-CLIP-170 antibody in blocking buffer at 1:1000 (4°C, overnight). For extreme sparsity, titrate to 1:5000.
- Wash 5x with PBS over 60 min.
- Secondary Antibody Incubation: Incubate with STED-optimized secondary antibody (e.g., Abberior STAR RED) at 1:400 (room temp, 45 min) in the dark.
- Wash 5x with PBS over 60 min.
- Post-fix with 4% PFA for 10 min to stabilize labeling.
- Mount in ProLong Glass antifade mountant. Cure for 48 hrs at RT before imaging.
Protocol 2.2: Gated STED Imaging for Cortical Optical Sectioning
Objective: To acquire super-resolution images of the cell cortex with enhanced SNR using time-gated detection. System Setup: Commercial or custom STED microscope with pulsed excitation (e.g., 590 nm), pulsed STED (775 nm), and time-correlated single photon counting (TCSPC) capability. Procedure:
- Locate cell cortex using confocal mode with low excitation power (≈1 µW).
- Switch to STED mode. Set STED laser to circular polarization and 40% of maximum power (≈80 MW/cm² at focal plane) as a starting point.
- Set TCSPC parameters: Excitation delay: 0 ns; Gating start: 0.5 ns; Gating width: 5.5 ns. Adjust based on fluorophore lifetime.
- Acquire a test STED image with a pixel size of 15 nm and a dwell time of 10 µs.
- Optimize Iteratively: a. If signal is weak, increase excitation power in 0.5 µW steps (max 10 µW). b. If background is high, increase gating start time in 0.2 ns steps. c. If resolution is poor, increase STED power in 5% steps, monitoring for photobleaching.
- Acquire z-stacks with a step size of 100 nm through the cortical region (≈1 µm total).
- Deconvolution: Process raw STED images with a Richardson-Lucy deconvolution algorithm using an experimentally measured STED point spread function (PSF) for final SNR enhancement.
Visualization Diagrams
Diagram Title: STED SNR Optimization Workflow for Cortical Imaging
Diagram Title: Microtubule-Cortex Connection & Signaling Pathway
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for SNR-Optimized Cortical STED
| Item | Function & Rationale | Example Product/Catalog # |
|---|---|---|
| STED-Optimized Fluorophore | High photostability, low triplet-state yield, and emission >600 nm to minimize cellular autofluorescence. Critical for clean signal. | Abberior STAR RED; Abberior STAR 635P |
| High-Affinity, Monoclonal Primary Antibodies | Ensures high specificity, reducing off-target labeling which contributes to background noise in dense regions. | Mouse anti-EB1 [clone 5]; Rabbit anti-CLASP2 |
| Sparse Labeling-Compatible Secondary | Conjugated to STED dye at a controlled, low F/P ratio to prevent signal overcrowding. | Abberior STAR RED F(ab)₂ Fragment, Anti-Mouse IgG (H+L) |
| RI-Matched Mounting Medium | Reduces light scattering and spherical aberration, maximizing signal collection efficiency at cortical depths. | ProLong Glass Antifade Mountant (Thermo Fisher, P36980) |
| High-Precision Coverslips #1.5H | Consistent thickness (170 µm ± 5 µm) is essential for maintaining STED doughnut shape and resolution. | Marienfeld Superior, #1.5H (0.17 mm) high-precision |
| STED Microscope with TCSPC | Hardware enabling time-gated detection to filter out early photon background from stimulated emission light. | Abberior Expert Line; Leica Stellaris 8 STED with TauSense |
| PSF Measurement Beads | Required for generating accurate PSF models for deconvolution, enhancing final SNR and resolution. | TetraSpeck Microspheres (0.1 µm, Thermo Fisher, T7279) |
In a thesis investigating microtubule-cortex connections using STED super-resolution microscopy, image fidelity is paramount. Precise visualization of proteins like CLASPs, dynein, or actin at these interfaces requires resolving structures at the sub-diffraction scale. Aberrations (optical imperfections) and misalignment (mechanical drift) in the STED beam directly degrade the effective resolution and signal-to-noise ratio, leading to artifactual data and erroneous biological conclusions. This application note provides detailed protocols for diagnosing, avoiding, and correcting these issues to ensure quantitative accuracy in cortical cytoskeleton research.
Quantitative Impact of Aberrations on STED Performance
Aberrations in the depletion beam (typically a donut or bottle beam) reduce its intensity at the zero point and distort its shape. This directly diminishes the achievable resolution, as defined by the STED resolution formula: d ≈ λ / (2 NA √(1 + I/Is)).
Table 1: Impact of Common Aberrations on STED Beam and Image Quality
| Aberration Type | Effect on Depletion Donut | Quantitative Impact on Resolution | Observed Artifact in Microtubule Images |
|---|---|---|---|
| Spherical (Defocus) | Blurred zero, reduced intensity at center. | Resolution can degrade by 30-50%. | Loss of fine detail; microtubules appear smeared. |
| Astigmatism | Donut becomes elliptical, zero point elongates. | Resolution anisotropic; can differ by 20-40% along axes. | Microtubules appear sharper in one direction than another. |
| Coma | Donut shape becomes asymmetric, zero shifts. | Local resolution loss (up to 60%) in peripheral FOV. | Microtubules near image edges are poorly resolved vs. center. |
| Sample-Induced (Refractive Index Mismatch) | Severe distortion and wandering of donut. | Resolution can degrade >70%, effective NA reduced. | Inconsistent resolution through depth in cortical samples. |
Protocols for Avoiding and Correcting Aberrations
Protocol 3.1: Daily Pre-Imaging Beam Alignment and Quality Check
Objective: Verify STED donut shape and co-alignment with the excitation beam. Materials: 100 nm crimson fluorescent beads, high-precision coverslip (#1.5H), immersion oil (matched to sample RI). Workflow:
- Prepare a sparse sample of beads on a coverslip.
- Switch to STED mode with depletion power set to a low level (e.g., 10%).
- Image a single bead with the detection pinhole fully open.
- Analyze the donut: The bead's emission pattern should show a clear, symmetric donut with a central zero. Use line profile analysis.
- Check co-alignment: The central zero of the donut must coincide with the peak of the excitation spot. Use software alignment tools to adjust the STED beam path if necessary.
- Quantify: Measure the intensity ratio between the donut peak and its center (should be >10:1 for a good beam).
Protocol 3.2: Active Wavefront Correction using a Deformable Mirror (DM)
Objective: Correct for system and sample-induced aberrations in real-time. Materials: Fluorescent guide star (bead or bright feature in sample), deformable mirror integrated into STED beam path, wavefront sensor (or sensorless algorithm). Workflow (Sensorless Approach):
- Focus on a bright, isolated reference structure (e.g., a bead or a dense tubulin cluster) within your cortical sample.
- Define a quality metric (e.g., total intensity in a confocal pinhole, or sharpness of the donut's inverse image).
- Command the DM to sequentially apply a set of predetermined aberration modes (Zernike polynomials).
- For each mode, measure the change in the quality metric.
- Use an optimization algorithm (e.g., steepest ascent) to determine the DM shape that maximizes the metric.
- Apply this correction to the depletion (and optionally, excitation) beam for subsequent imaging of the region.
Diagram Title: Sensorless Adaptive Optics Workflow for STED (82 chars)
Protocol 3.3: Post-Processing Deconvolution for Residual Blur
Objective: Enhance final image quality by computationally compensating for a measured or estimated effective PSF. Materials: Raw STED image stack, experimentally acquired or calculated STED PSF (or a robust theoretical model). Workflow:
- PSF Estimation: Ideally, measure the in-situ PSF using a 100 nm bead embedded in your sample hydrogel. Alternatively, use a theoretically simulated STED PSF incorporating your measured donut parameters.
- Pre-processing: Apply mild background subtraction and noise reduction to the raw data.
- Deconvolution: Use a constrained iterative algorithm (e.g., Richardson-Lucy, Huygens software) with the estimated STED PSF.
- Validation: Compare deconvolved microtubule line profiles with the raw data; resolution (FWHM) should improve without introducing ringing artifacts.
The Scientist's Toolkit: Key Reagents and Materials
Table 2: Essential Research Reagent Solutions for STED Alignment & Microtubule Imaging
| Item | Function & Rationale |
|---|---|
| 100 nm Crimson Fluorescent Beads (e.g., Dark Red) | Gold standard for PSF measurement. Far-red emission minimizes cross-talk, ideal for checking STED donut shape and alignment. |
| #1.5 High-Precision Coverslips (170 ± 5 µm) | Critical for minimizing spherical aberration. Thickness tolerance ensures optimal performance of oil immersion objectives. |
| Index-Matched Immersion Oil & Mounting Medium | Reduces sample-induced aberrations. For live-cell cortical imaging, choose media matching cytoplasmic RI (~1.38). |
| Fiducial Markers (e.g., TetraSpeck beads) | Multi-color beads for assessing and correcting channel registration in multi-color STED experiments of microtubule-actin overlap. |
| Anti-Fade Mounting Media (for fixed samples) | Preserves fluorophore brightness under intense STED illumination. Essential for reproducible imaging of fixed microtubule networks. |
| Deformable Mirror Kit (e.g., ALPAO, BMC) | Hardware for active wavefront correction. Integrated into beam path to compensate for system and sample aberrations dynamically. |
| STED-optimized Primary Antibodies & Labels | Use small, bright, photostable dyes (e.g., Abberior STAR, Alexa Fluor 594) conjugated directly to antibodies for labeling cortical targets. |
Strategies for Multi-Color STED Imaging of Microtubules and Cortical Markers
Within the broader thesis investigating microtubule-cortex connections, the application of super-resolution microscopy is indispensable. Stimulated Emission Depletion (STED) microscopy overcomes the diffraction limit, enabling the visualization of the precise spatial relationship between dynamic microtubules and the static cortical actin meshwork or membrane-associated proteins. Successful multi-color STED imaging of these distinct targets presents significant challenges in spectral separation, photostability, and sample preparation. These application notes provide current strategies and detailed protocols to address these challenges.
Key Considerations for Multi-Color STED
The primary challenge lies in selecting fluorophore pairs with well-separated emission spectra that can be efficiently depleted by the available STED lasers (typically 592 nm, 660 nm, or 775 nm). Photostability under high-intensity STED depletion is paramount. Furthermore, sample fixation and labeling must preserve ultrastructure while allowing efficient antibody penetration.
Table 1: Recommended Fluorophore Pairs for 2-Color STED
| Target 1 (Microtubules) | Target 2 (Cortical Marker) | Excitation (nm) | STED Depletion (nm) | Emission Detection (nm) | Key Advantage |
|---|---|---|---|---|---|
| Abberior STAR RED | Abberior STAR ORANGE | 640 / 561 | 775 / 660 | 660-720 / 580-630 | Excellent separation, high photostability |
| ATTO 594 | KK 114 | 594 / 647 | 660 / 775 | 610-630 / 670-720 | Classic, well-characterized pair |
| Alexa Fluor 594 | Abberior STAR 635P | 594 / 635 | 775 / 775 | 610-630 / 650-720 | Common secondary antibodies, single STED line |
Table 2: Quantitative Performance Comparison
| Fluorophore Pair | Achievable Resolution (FWHM) | Relative Photostability (Frames before bleach) | Recommended Mounting Medium |
|---|---|---|---|
| STAR RED / STAR ORANGE | ~50 nm (Red), ~60 nm (Orange) | > 200 frames | ProLong Diamond / TRIS-Buffer + O₂ scavenger |
| ATTO 594 / KK 114 | ~60 nm (Green), ~70 nm (Red) | ~150 frames | Mowiol with NPG / GLOX |
| Alexa Fluor 594 / STAR635P | ~70 nm (Green), ~50 nm (Red) | ~120 frames | ProLong Glass |
Detailed Experimental Protocols
Protocol 1: Sample Preparation for Microtubule and Cortical Actin Preservation
- Cell Culture & Fixation: Grow cells on high-precision #1.5H coverslips. Fix with 4% formaldehyde + 0.1% glutaraldehyde in PEM buffer (100 mM PIPES, 5 mM EGTA, 2 mM MgCl₂, pH 6.8) for 10 min at 37°C.
- Quenching & Permeabilization: Quench autofluorescence with 0.1% NaBH₄ for 7 min. Permeabilize with 0.1% Triton X-100 in PBS for 5 min.
- Blocking: Block with 3% BSA + 0.05% Tween-20 in PBS for 1 hour.
- Immunostaining:
- Apply primary antibodies: mouse anti-α-tubulin (1:200) and rabbit anti-beta-II spectrin or anti-Ezrin (1:100) in blocking buffer overnight at 4°C.
- Wash 3x with PBS.
- Apply secondary antibodies conjugated to STED-optimized fluorophores (e.g., anti-mouse STAR RED, anti-rabbit STAR ORANGE) at 1:200 for 1 hour at RT. Perform this step in darkness.
- Post-Fixation: Fix again with 2.5% formaldehyde for 10 min to stabilize labeling.
- Mounting: Mount in ProLong Diamond Antifade mountant. Seal with nail polish. Store at 4°C in darkness.
Protocol 2: Two-Color STED Imaging Acquisition Parameters
This protocol assumes a gated-STED system with 592 nm and 660 nm depletion lasers.
- System Alignment: Prior to experiment, align the STED depletion donuts for both channels using 40 nm gold beads to ensure co-alignment.
- Sequential Acquisition Setup: Acquire channels sequentially to avoid cross-talk.
- Channel 1 (Microtubules - STAR RED): Excitation: 640 nm (pulsed), Depletion: 775 nm, Detection: 650-720 nm, Gating: 0.5-6 ns.
- Channel 2 (Cortex - STAR ORANGE): Excitation: 561 nm (pulsed), Depletion: 660 nm, Detection: 570-620 nm, Gating: 0.3-6 ns.
- Power Optimization: Start with low STED power (~10% of max) and increase until resolution improvement plateaus, monitoring for bleaching. Typical final powers: 5-15 µW (excitation), 10-30 mW (depletion) at sample plane.
- Pixel Size & Scan Speed: Set pixel size to 15-20 nm (1/3 of desired resolution). Use a scan speed of 400-800 lines per second with 4-8 line accumulations.
- Deconvolution: Apply a 2-3 iteration Huygens deconvolution using a measured STED PSF for optimal final resolution.
Visualization of Workflows and Relationships
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Multi-Color STED Imaging
| Item / Reagent | Function & Rationale | Example Product / Specification |
|---|---|---|
| STED-Optimized Fluorophores | High photostability and specific spectral properties for efficient depletion. | Abberior STAR dyes, ATTO dyes, KK114 |
| High-Precision Coverslips | Ensure optimal point spread function and imaging stability. | #1.5H, thickness 170 µm ± 5 µm |
| Crosslinking Fixative Mix | Preserves ultrastructure; low % glutaraldehyde improves MT fixation without excessive autofluorescence. | 4% Formaldehyde + 0.1-0.5% Glutaraldehyde |
| O₂ Scavenging Mountant | Reduces photobleaching during STED imaging by removing oxygen. | GLOX system (Glucose Oxidase + Catalase) or commercial ProLong Diamond |
| STED-Tuned Secondary Antibodies | Direct conjugation to optimal dyes ensures bright, specific labeling. | Anti-species IgG conjugated to STAR RED/ORANGE |
| Primary Antibodies (Monoclonal) | High specificity and affinity for targets like tubulin and cortical markers. | Clone DM1A (α-tubulin), Clone 42 (β-II Spectrin) |
| Alignment Nanobeads | Critical for aligning and calibrating the STED depletion donut. | 40 nm Gold Nanoparticles or Crimson beads |
Within the broader thesis investigating the nanoscale organization and dynamic interactions between microtubules and the actin cortex in cellular mechanobiology, STED (Stimulated Emission Depletion) microscopy is indispensable. This super-resolution technique resolves structures below the diffraction limit, crucial for visualizing how microtubule plus-ends engage with cortical actin networks during processes like cell migration and division. However, STED data is inherently affected by background noise, photon counting effects, and the residual depletion donut, necessitating robust deconvolution and processing for faithful image restoration and quantitative analysis. This document outlines best practices and protocols for this restoration pipeline.
Core Principles of STED Image Deconvolution
STED deconvolution aims to reverse the blurring introduced by the effective point spread function (PSF), which is smaller than in confocal microscopy but must be precisely characterized. Key considerations include:
- PSF Determination: The effective STED PSF must be measured experimentally using sub-resolution beads under identical imaging conditions (wavelength, power, depletion efficiency) as the biological sample.
- Noise Modeling: STED images, especially time-gated (gSTED), are dominated by Poisson (photon shot) noise. Deconvolution algorithms must incorporate this statistical model.
- Regularization: To prevent noise amplification, regularization parameters (e.g., Tikhonov, Total Variation) are essential to constrain the solution based on prior knowledge (e.g., non-negativity, signal sparsity).
Table 1: Comparison of Common Deconvolution Algorithms for STED
| Algorithm | Principle | Pros for STED | Cons for STED | Recommended Use Case |
|---|---|---|---|---|
| Classic Maximum Likelihood Estimation (MLE) | Iterative, models Poisson noise. | Good for photon-limited data. Simple. | Can amplify noise; slow convergence. | Initial processing, moderate signal-to-noise ratio (SNR) data. |
| Richardson-Lucy with Total Variation (RL-TV) | MLE with TV regularization promoting smoothness. | Suppresses noise; preserves edges. | Risk of over-smoothing fine details. | Noisy datasets where cortical structures are relatively coarse. |
| DeconvolutionLab2 (Iterative Deconvolve) | Modular, offers several algorithms (e.g., Tikhonov-Miller). | Highly configurable; benchmarked. | Requires parameter tuning expertise. | General use, especially with known experimental PSF. |
| Sparse Deconvolution (e.g., Huygens) | Assumes signal is sparse in a transform domain. | Excellent for resolving dense structures; high resolution gain. | Computationally intensive; may produce artifacts. | Dense microtubule arrays near the cortex. |
| Machine Learning-Based (e.g., CARE, Content-Aware) | Trained network restores images using examples. | Powerful for extreme low-light or fast imaging. | Requires large, high-quality training datasets. | Live-cell STED of dynamic cortex-microtubule interactions. |
Experimental Protocol: STED Imaging and Restoration of Microtubule Cortex Proximity
Sample Preparation (Fixed Cells)
- Cell Culture: Plate human osteosarcoma (U2OS) cells on #1.5 high-precision coverslips.
- Fixation and Permeabilization: At 37°C, fix with 4% paraformaldehyde (PFA) + 0.1% glutaraldehyde in PEM buffer (100 mM PIPES, 1 mM EGTA, 1 mM MgCl₂, pH 6.8) for 10 min. Quench with 0.1% NaBH₄. Permeabilize with 0.5% Triton X-100.
- Immunostaining:
- Block with 3% BSA in PBS for 1 hr.
- Incubate with primary antibodies: mouse anti-α-tubulin (1:500) and rabbit anti-ERM (Ezrin/Radixin/Moesin, 1:400, as cortex marker) overnight at 4°C.
- Wash 3x with PBS.
- Incubate with secondary antibodies: STAR RED for tubulin (ex/em: ~640/660nm) and STAR ORANGE for ERM (ex/em: ~560/585nm) coupled to Abberior STED dyes. Use Atto 594 for tubulin if two-color STED with 775nm depletion is planned. Incubate for 1 hr at RT.
- Wash and mount in ProLong Glass antifade mountant.
STED Microscopy Acquisition
- Instrument: Use a commercial gated-STED system (e.g., Abberior, Leica, or Zeiss) equipped with a 775 nm depletion laser and time-gated detection.
- Acquisition Settings:
- Confocal Reference: First, acquire a confocal stack (pinhole: 1 Airy unit) for both channels.
- STED Imaging: Activate the STED depletion laser (775 nm). Set the gating delay to 0.5–1.5 ns and width to 5–7 ns to suppress background fluorescence.
- Pixel Size: 20 nm (well below the expected STED resolution of ~50 nm).
- Dwell Time: 10-20 µs/pixel to ensure sufficient photon count.
- Laser Powers: Optimize to maximize signal and minimize photobleaching. Typical: Excitation (580 nm/640 nm) at 1-5% of saturating power; Depletion (775 nm) at 20-40 mW at the sample.
- PSF Measurement: Image 40 nm crimson fluorescent beads under identical conditions to generate the experimental PSF for deconvolution.
Image Restoration Workflow Protocol
- Pre-processing (in Fiji/ImageJ):
- Apply a Gaussian blur (σ = 0.5 px) to the raw STED image to suppress single-pixel noise.
- Perform flat-field correction if illumination is uneven.
- Subtract background using a "rolling ball" algorithm (radius: 5-10 pixels).
- Deconvolution (using Huygens Software or DeconvolutionLab2):
- Load Data: Import the raw STED image stack and the measured PSF image.
- Set Parameters: Signal-to-noise ratio (SNR): Estimate from background (start with 20). Iterations: 40-60. Use "Classic MLE" or "Quick MLE" algorithm.
- Regularization: Enable "Tikhonov-Miller" regularization with a medium strength setting.
- Execute: Run deconvolution. Check for noise amplification and adjust regularization if needed.
- Post-processing & Analysis:
- Visualization: Apply a linear contrast stretch for display.
- Colocalization Analysis: For the deconvolved two-channel image, use the JACoP plugin in Fiji to calculate Manders' overlap coefficients (M1, M2) between microtubule and cortical ERM signals within a 200 nm proximity band from the plasma membrane.
- Distance Measurement: Use the "Line Scan" tool to plot intensity profiles perpendicular to the cell edge and measure the distance from microtubule ends to peak cortical signal.
Visualization of Workflows and Relationships
STED Image Restoration Pipeline
From Thesis Goal to Biological Insight
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for STED of Microtubule-Cortex Connections
| Item / Reagent | Function in Experiment | Key Specification / Note |
|---|---|---|
| STED-Compatible Dyes (e.g., Abberior STAR RED/ORANGE, ATTO 594) | High photon output and photostability under STED depletion laser. | Must match available excitation/depletion lasers (e.g., 590/640 nm ex, 775 nm depletion). |
| High-Precision Coverslips (#1.5H, 0.17mm ± 0.005mm) | Ensure optimal objective correction and minimal spherical aberration. | Critical for achieving maximal resolution. |
| Anti-fade Mountant (e.g., ProLong Glass, Mowiol with antifade) | Preserve fluorescence signal during STED scanning. | "ProLong Glass" hardens for stable imaging plane. |
| Primary Antibodies: anti-α-tubulin, anti-ERM (ezrin) | Specific labeling of microtubules and the actin cortex. | Validate for super-resolution; monoclonal antibodies preferred. |
| Fluorescent Nanobeads (40nm, crimson/dark red) | Experimental measurement of the system's Point Spread Function (PSF). | Essential for accurate, sample-specific deconvolution. |
| Image Deconvolution Software (e.g., Huygens, DeconvolutionLab2) | Algorithmic restoration of super-resolution images. | Must support experimental PSF and Poisson noise modeling. |
| Analysis Software (e.g., Fiji with JACoP, Imaris) | Quantitative analysis of colocalization and nanoscale distances. | Enables extraction of thesis-relevant metrics from restored images. |
Benchmarking STED: How It Stacks Up Against Other Super-Resolution Methods for Cytoskeletal Imaging
This application note directly compares Stimulated Emission Depletion (STED) and Photoactivated Localization Microscopy (PALM)/Stochastic Optical Reconstruction Microscopy (STORM) within the specific research framework of investigating nanoscale connections between microtubules and the actin cortex. Understanding the precise architecture and dynamic interface of these cytoskeletal systems is crucial for elucidating mechanisms in cell division, migration, and intracellular transport—key targets in oncological and neurological drug development. The choice of super-resolution technique profoundly impacts the ability to resolve these structures, quantify labeling density, and perform live-cell imaging.
Quantitative Comparison Table
| Parameter | STED Microscopy | PALM / STORM |
|---|---|---|
| Fundamental Principle | Deterministic switching via depleting doughnut. | Stochastic switching & single-molecule localization. |
| Practical Resolution | 30-80 nm laterally. | 10-30 nm laterally. |
| Temporal Resolution | High (Video-rate possible). Suitable for live-cell dynamics. | Low (Seconds to minutes per frame). Primarily for fixed cells. |
| Labeling Density Requirement | High. Requires dense, uniform labeling for continuous structures. | Low. Relies on sparse, stochastic activation. |
| Probe Compatibility | Conventional fluorescent dyes (e.g., Atto 594, Alexa Fluor 647) and photoswitchable proteins. | Special photoswitchable/photoactivatable dyes (e.g., Alexa Fluor 647 paired with imaging buffer) or proteins (mEos, Dendra). |
| Multicolor Imaging | Relatively straightforward, similar to confocal. | Challenging due to cross-talk in activation/bleaching; requires careful sequential imaging. |
| Sample Preparation | Similar to confocal; often simpler buffer requirements. | Can require specific oxygen-scavenging/thiol imaging buffers for optimal blinking. |
| Primary Application in Microtubule-Cortex Studies | Live-cell nanodynamics: Imaging cortex-proximal microtubule ends, trafficking events. | Ultra-structural mapping: Nanoscale architecture of cortical anchoring complexes in fixed cells. |
Detailed Experimental Protocols
Protocol A: STED Imaging of Microtubule Tips at the Cortex in Live Cells
Objective: To visualize the dynamic interaction of microtubule plus-ends with the subcortical actin meshwork in live COS-7 cells.
Reagents & Materials: See "Scientist's Toolkit" below.
Procedure:
- Cell Preparation: Plate COS-7 cells on high-performance coverslips. Transfect with expression constructs for EB3 (microtubule plus-end binding protein) tagged with a bright, photostable dye compatible with STED (e.g., SNAP-tag, then label with SNAP-Cell 647).
- Cortex Staining (Optional): Incubate live cells with SiR-actin (0.5 µM) for 1 hour to label the actin cortex. Use confocal channel for actin to avoid STED depletion laser cross-talk.
- Microscope Setup: Mount sample in live-cell imaging chamber (37°C, 5% CO₂). On a gated-STED system:
- Configure 640 nm excitation laser and 775 nm depletion (doughnut) laser.
- Set detection gate to 0.8-6 ns to suppress background.
- Use a 100x/NA 1.4 oil immersion objective.
- Acquisition: Acquire time-series STED images (512x512 pixels) at 2-second intervals for 2 minutes. Use pixel sizes of 20-30 nm. Keep STED laser power as low as possible (e.g., 20-40% of max) to minimize phototoxicity.
- Analysis: Use kymograph analysis along cell edges to track EB3-comet trajectories and pause events at the cortex.
Protocol B: PALM/STORM Reconstruction of Cortical Microtubule Anchoring Sites
Objective: To achieve ultrastructural resolution of microtubule ends embedded within the cortical actin network in fixed RPE-1 cells.
Reagents & Materials: See "Scientist's Toolkit" below.
Procedure:
- Sample Fixation & Immunostaining: Fix cells with 4% PFA + 0.1% glutaraldehyde for 10 min. Quench with 0.1% NaBH₄. Permeabilize and block. Incubate with primary antibody against α-tubulin and then with secondary antibody conjugated to photoswitchable dye (e.g., Alexa Fluor 647).
- PALM/STORM Imaging Buffer Preparation: Prepare a oxygen-scavenging, thiol-containing buffer: 50 mM Tris, 10 mM NaCl, 10% Glucose, 0.5 mg/mL Glucose Oxidase, 40 µg/mL Catalase, and 10-100 mM β-Mercaptoethylamine (MEA). Adjust pH to 8.0.
- Sample Mounting: Apply 100-200 µL of imaging buffer to the sample and seal with a clean coverslip using nail polish.
- Microscope Setup: On a TIRF/PALM system equipped with 405 nm activation and 642 nm excitation lasers:
- Use a 100x/NA 1.49 oil immersion TIRF objective.
- Set the 642 nm laser to high power (~2-5 kW/cm²) to drive Alexa Fluor 647 into a dark state.
- Use low power 405 nm laser to stochastically reactivate molecules.
- Acquisition: Capture 15,000-30,000 frames at an exposure time of 20-30 ms per frame. Ensure localization density is sparse in each frame.
- Reconstruction & Analysis: Use localization software (e.g., ThunderSTORM, Picasso) to fit single-molecule positions and render a super-resolution image. Co-align with a confocal image of phalloidin-stained actin to correlate microtubule ends with cortical actin density.
Visualization Diagrams
Title: Technique Selection Workflow for Microtubule-Cortex Imaging
Title: STED Principle: PSF Reduction via Depletion
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Experiment | Example Product/Catalog # |
|---|---|---|
| SNAP-Cell 647 | Covalent, bright, and photostable label for live-cell STED of tagged proteins (e.g., EB3). | New England Biolabs, #S9106S |
| SiR-Actin / SiR-Tubulin | Live-cell compatible, far-red fluorescent probes for staining cytoskeleton with minimal toxicity. | Cytoskeleton, Inc., #CY-SC001 / #CY-SC002 |
| Alexa Fluor 647 NHS Ester | Photoswitchable dye for antibody conjugation in fixed-sample PALM/STORM. | Thermo Fisher Scientific, #A37573 |
| Glucose Oxidase Catalase System | Critical components of PALM/STORM imaging buffer to reduce oxygen and photobleaching. | Sigma-Aldrich, #G2133 / #C40 |
| β-Mercaptoethylamine (MEA) | Thiol-based switching/blinking enhancer in PALM/STORM buffer for Alexa Fluor 647. | Sigma-Aldrich, #30070 |
| Mounting Media for STED | Anti-fade, refractive-index matched media for fixed samples. | e.g., ProLong Diamond |
Investigating the dynamic interface between microtubules and the actin cortex is fundamental to understanding cell division, migration, and polarity. These processes involve rapid, nanoscale rearrangements requiring live-cell imaging with high spatial and temporal resolution. This application note, framed within a thesis on STED microscopy for this research, compares two leading super-resolution techniques—Stimulated Emission Depletion (STED) and Structured Illumination Microscopy (SIM)—specifically for live-cell imaging of microtubule-cortex interactions, focusing on speed, ease of use, and practical implementation.
Technology Comparison: STED vs. SIM for Live-Cell Imaging
A current comparison of key performance parameters for live-cell imaging is summarized below.
Table 1: Quantitative Comparison of STED and SIM for Live-Cell Imaging
| Parameter | STED (Gated, Live-Cell Optimized) | SIM (TIRF or Lattice-based, Live-Cell) |
|---|---|---|
| Spatial Resolution (XY) | 50-80 nm | 90-120 nm |
| Temporal Resolution (Frame Rate) | 1-5 seconds per frame (for a 512x512 px field) | 0.1-1 second per frame (for a 512x512 px field) |
| Typical Excitation Power | Moderate-High (depletion laser adds intensity) | Low-Moderate |
| Photobleaching/Phototoxicity | Higher (due to intense, focused beams) | Lower |
| Ease of Sample Preparation | Requires special high-performance dyes (e.g., Abberior STAR, ATTO dyes) | Compatible with many standard fluorescent proteins and dyes |
| Ease of Operation | More complex; requires precise laser alignment and photon gating optimization. | More straightforward; commercial systems offer automated reconstruction. |
| Maximum Imaging Depth | ~5-10 µm (best near coverslip) | ~20-50 µm (with 3D-SIM) |
| Data Processing | Direct image acquisition; deconvolution optional. | Mandatory computational reconstruction, which can introduce artifacts. |
Detailed Application Protocols
Protocol 1: Live-Cell STED Imaging of Cortical Microtubule Tips
Objective: Image the plus-end dynamics of microtubules interacting with the cell cortex in live COS-7 cells. Reagent Solutions: See Table 2.
Method:
- Cell Preparation: Plate COS-7 cells on high-performance #1.5H glass-bottom dishes. Transfect with a plasmid encoding EB3 (microtubule plus-end binding protein) tagged with Abberior STAR ORANGE or Janelia Fluor 549 using a suitable transfection reagent.
- Dye Selection & Imaging Buffer: Use live-cell imaging medium (e.g., FluoroBrite DMEM) supplemented with 10% FBS and 25mM HEPES. For optimal STED performance, avoid phenol red.
- Microscope Setup (STED):
- Excitation: Use a 560 nm pulsed laser line.
- Depletion: Use a 775 nm STED donut beam.
- Detection: Use a gated detector (time-gated detection recommended, e.g., 0.5-6 ns delay) to suppress background emission.
- Scanning Parameters: Set pixel size to 20 nm, pixel dwell time to 5 µs. Use bidirectional scanning. For a 10 µm x 10 µm field, this yields a frame time of ~1.3 seconds.
- Live Imaging: Maintain cells at 37°C and 5% CO₂. Acquire time-series images at 2-second intervals for 2-5 minutes.
- Analysis: Use software (e.g., ImageJ with TrackMate or commercial packages) to track EB3 comets. Measure dwell time and distance of comets at the cell periphery as a proxy for cortex interaction.
Protocol 2: Live-Cell TIRF-SIM Imaging of Actin-Microtubule Proximity
Objective: Visualize co-alignment of actin filaments and microtubules at the basal cortex. Reagent Solutions: See Table 2.
Method:
- Cell Preparation: Plate NIH/3T3 cells on glass-bottom dishes. Co-transfect with plasmids for Lifeact-mNeonGreen (actin label) and mRuby3-Tubulin (microtubule label).
- Microscope Setup (TIRF-SIM):
- Excitation: Use 488 nm and 561 nm laser lines.
- TIRF Angle: Calibrate to achieve a sub-200 nm evanescent field.
- SIM Pattern Generation: Use a diffraction grating (in commercial systems) to project structured illumination patterns (typically 3 phases and 3 rotations).
- Acquisition: System automatically acquires 9 raw images per color per time point.
- Live Imaging: Maintain physiological conditions. Acquire a time series every 5 seconds. A full SR reconstruction per time point may take ~0.5-1 second with GPU-accelerated software.
- Image Reconstruction: Use the microscope manufacturer's software (e.g., ZEISS ZEN, Nikon NIS-Elements) to reconstruct super-resolved images. Ensure the correct pattern period and modulation contrast parameters are set.
- Co-Localization Analysis: Reconstructed channels are aligned. Use correlation analysis (e.g., Pearson's coefficient in a defined cortical region of interest) or line profile analysis to assess spatial relationships.
Visualization Diagrams
Live-Cell STED Imaging Workflow
Live-Cell SIM Imaging & Processing Workflow
STED vs SIM Live-Cell Selection Guide
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Live-Cell Super-Resolution of Cytoskeleton
| Item | Function & Rationale | Example Product/Brand |
|---|---|---|
| High-Performance STED Dyes | Fluorophores with high photostability and emission compatible with STED depletion lasers. Critical for STED live-cell imaging. | Abberior STAR RED/ORANGE; Janelia Fluor 549, 646; ATTO 590, 647 |
| Standard Fluorescent Proteins | Bright, photostable FPs compatible with SIM illumination patterns. Enable multi-color live SIM. | mNeonGreen, mRuby3, mScarlet |
| Live-Cell Imaging Medium | Phenol-red free medium with buffering to maintain pH without CO₂. Reduces background. | FluoroBrite DMEM, CO₂-Independent Medium |
| #1.5H Coverslips/Dishes | High-precision glass (170 µm ± 5 µm thickness) essential for maintaining optical integrity, especially for TIRF-SIM. | MatTek dishes, Ibidi µ-Dish |
| Fiducial Markers for Drift Correction | Gold or fluorescent nanoparticles for sub-pixel stabilization during long time-lapses. | TetraSpeck microspheres, Gold nanoparticles (100 nm) |
| Microtubule Plus-End Marker | Protein to label growing microtubule ends for dynamic studies. | EB3, Clip-170 (tagged with suitable FP/dye) |
| Actin Cortex Marker | Protein to label filamentous actin without disrupting dynamics. | Lifeact, Utrophin calponin homology domain (tagged) |
| Transfection Reagent | For efficient delivery of plasmid DNA encoding fluorescent constructs. | Lipofectamine 3000, FuGENE HD, jetPRIME |
Within the context of a thesis on STED super-resolution microscopy for investigating microtubule-cortex connections, this application note details quantitative analysis of molecular distances and colocalization at the cell cortex. Precise measurement of the spatial relationship between microtubule plus-ends, cortical actin, and associated protein complexes is crucial for understanding mechanisms of cell division, migration, and signaling.
Quantitative Data on Cortex-Associated Proteins
Summary of typical distances and colocalization metrics for key components at the microtubule-cortex interface, as measured by STED microscopy.
Table 1: Characteristic Distances at the Cell Cortex
| Protein/Pair | Average Distance (nm) ± SD | Biological Context | Measurement Method |
|---|---|---|---|
| EB1 (MT tip) to Cell Membrane | 52 ± 18 | Leading Edge, Migrating Cell | Tip-to-Edge Distance |
| CLASP to Actin Cortex | < 80 | Interphase Cortex | Nearest Neighbor Distance |
| Dystrophin-related complex to MT tip | 120 ± 40 | Muscle Cell Cortex | Centroid-to-Centroid |
| Septin ring to submembrane actin | 90 ± 25 | Cytokinetic Furrow | Line Profile FWHM |
Table 2: Colocalization Analysis of Cortical Complexes
| Protein A | Protein B | Pearson's Coefficient (Range) | Manders' Overlap Coefficients (M1/M2) | Biological System |
|---|---|---|---|---|
| CLIP-170 | EB1 | 0.65 - 0.85 | 0.72 / 0.81 | Fibroblast Lamella |
| Dynactin (p150) | CLASP2 | 0.55 - 0.75 | 0.68 / 0.61 | Neuronal Growth Cone |
| Actin (Phalloidin) | Cortical MT | 0.10 - 0.30 | 0.15 / 0.45 | Epithelial Cell Cortex |
Experimental Protocols
Protocol 2.1: Sample Preparation for Cortical STED Imaging
- Primary Fixation: Culture cells on high-precision #1.5H glass coverslips. Fix with 4% PFA + 0.1% glutaraldehyde in PEM buffer (100 mM PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.8) for 10 min at 37°C.
- Quenching & Permeabilization: Quench autofluorescence with 0.1% NaBH4 for 7 min. Permeabilize with 0.2% Triton X-100 for 10 min.
- Immunostaining: Incubate with primary antibodies (e.g., anti-α-Tubulin, anti-EB1, anti-p34/Arp11) diluted in 3% BSA/PBS overnight at 4°C. Use secondary antibodies conjugated with STAR RED, ATTO 590, or Abberior STAR ORANGE for STED compatibility.
- Mounting: Mount in commercially available STED mounting medium (e.g., Abberior Mount Solid Antifade) to minimize drift and preserve fluorescence.
Protocol 2.2: STED Image Acquisition for Distance Analysis
- System Setup: Use a gated STED microscope (e.g., Leica SP8 STED, Abberior FACILITY) with a 775 nm depletion laser. Set detection gates to 0.3-6 ns to suppress background.
- Calibration: Image 40 nm crimson fluorescent beads to determine the effective resolution (typically 30-50 nm lateral).
- Acquisition Parameters: Acquire sequential STED channels. Use a 100x/1.4 NA oil objective. Pixel size: 10-20 nm; line accumulation: 8-12; pixel dwell time: 5-10 µs. Maintain depletion laser power at the minimum required for desired resolution to limit photobleaching.
- Region Selection: Acquire z-stacks (3-5 slices, 200 nm step) focusing on the basal cell cortex adherent to the coverslip.
Protocol 2.3: Quantitative Analysis of Distances & Colocalization
- A. Preprocessing:
- Deconvolve images using Huygens Professional with a calculated PSF.
- Apply a mild Gaussian filter (σ = 1 pixel) to reduce noise.
- Create a binary mask of the cell cortex using the actin channel.
- B. Distance Measurement (Tip-to-Cortex):
- Identify microtubule plus-ends using the EB1 channel with a local maximum detection algorithm (e.g., TrackMate in Fiji).
- Detect the cortical edge from the actin or membrane stain using a sub-pixel edge detection algorithm (e.g.,
findEdgesin Fiji). - For each EB1 point, calculate the shortest Euclidean distance to the cortical edge. Export distances for statistical analysis.
- C. Colocalization Analysis (Within Cortex Region of Interest):
- Apply the cortical mask to both channels of interest.
- Calculate Pearson's Correlation Coefficient (PCC) using the JaCoP plugin in Fiji. PCC assesses linear correlation of pixel intensities.
- Calculate Manders' Overlap Coefficients (M1 & M2) using JaCoP with Costes' automatic thresholding. M1 represents the fraction of Protein A colocalizing with Protein B, and vice versa for M2.
- Perform statistical significance testing using Costes' randomization approach (100 iterations).
Visualized Workflows & Pathways
STED Cortical Analysis Workflow
Microtubule-Cortex Connection Pathway
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for STED Cortical Analysis
| Item | Function & Critical Feature |
|---|---|
| STED-Compatible Fluorophores (e.g., Abberior STAR RED, ATTO 590) | High photostability and brightness under STED depletion laser; essential for sustained super-resolution imaging. |
| High-Precision Coverslips (#1.5H, 170 ± 5 µm) | Optimal thickness for oil immersion objectives; ensures minimal spherical aberration. |
| STED Optimized Mounting Medium (e.g., ProLong Diamond, Abberior Mount) | Preserves fluorescence, reduces photobleaching, and minimizes sample drift during acquisition. |
| Validated Primary Antibodies (e.g., anti-α-Tubulin [DM1A], anti-EB1 [5EB11]) | High specificity and affinity for target antigen; validation for fixed-cell super-resolution is crucial. |
| Cortex-Specific Probes (e.g., Lifeact-mRuby3, Membrane GFP) | Genetically encoded markers for live-cell cortical actin or membrane prior to fixation. |
| 40 nm Crimson Fluorescent Beads | Essential for daily point spread function (PSF) measurement and system resolution calibration. |
| Gated Detection STED Microscope (e.g., Leica TCS SP8 STED 3X) | Provides the <50 nm resolution necessary to resolve molecular-scale distances at the cortex. |
Application Notes
Within the broader thesis investigating microtubule-cortex connections, validating super-resolution STED microscopy findings with electron microscopy (EM) is paramount. STED provides nanoscale resolution (~30-50 nm) of fluorescently labeled proteins in a near-native state but lacks the ultrastructural context. EM offers unparalleled resolution (<5 nm) of cellular architecture but is limited in molecular specificity. Correlative Light and Electron Microscopy (CLEM) bridges this gap, allowing precise registration of STED-localized proteins, such as cortical actin regulators or microtubule minus-end-binding proteins, with their ultrastructural environment at the cell periphery.
Key quantitative advantages of this correlation are summarized below:
Table 1: Quantitative Comparison of STED and EM Modalities
| Parameter | STED Microscopy | Transmission Electron Microscopy (TEM) | Correlative STED-EM Outcome |
|---|---|---|---|
| Lateral Resolution | 30-50 nm | 0.2-5 nm | EM validates STED-measured distances between cortical markers & microtubule ends. |
| Axial Resolution | 500-700 nm | ~5 nm (for 70 nm sections) | Confirms 3D spatial relationships at cortex. |
| Molecular Specificity | High (via fluorescent tags) | Low (requires immunogold) | Direct correlation of protein presence with ultrastructure. |
| Sample Preparation | Live or fixed, hydrated | Fixed, dehydrated, resin-embedded | Requires compatible protocols (e.g., Tokuyasu cryosections). |
| Typical Label Size | ~5-15 nm (organic dye) | 5-15 nm (colloidal gold particle) | Enables co-localization analysis. |
| Accuracy of Correlation | N/A | N/A | Registration precision of 50-100 nm achievable with fiducial markers. |
Table 2: Typical Correlation Metrics for Microtubule-Cortex Analysis
| Measured Feature | STED Measurement (Mean ± SD) | EM Validation Measurement | Biological Insight Gained |
|---|---|---|---|
| Distance from microtubule end to plasma membrane | 150 ± 40 nm | 145 ± 30 nm | Validates cortical microtubule capture distance. |
| Diameter of cortical actin mesh | 35 ± 15 nm (via label size) | 25 ± 5 nm (direct visualization) | Confirms STED can resolve dense cortical networks. |
| Co-localization of protein X with microtubule end | 75% of puncta within 100 nm | Immunogold particles within 50 nm of end | Confirms protein's role in microtubule anchoring. |
Detailed Experimental Protocols
Protocol 1: Sample Preparation for Correlative STED and TEM on Adherent Cells Objective: Prepare biological samples (e.g., RPE-1 or HeLa cells) expressing fluorescently tagged proteins of interest (e.g., CLASP2-GFP) for sequential STED imaging and TEM ultrastructure analysis.
- Cell Culture and Fixation: Grow cells on #1.5 glass-bottom dishes with etched finder grids. Treat with drugs if required (e.g., nocodazole for microtubule depolymerization). Fix with 4% paraformaldehyde (PFA) and 0.1% glutaraldehyde (GA) in PBS for 15 min at 37°C.
- Immunostaining for STED: Quench autofluorescence with 0.1% sodium borohydride. Permeabilize with 0.1% Triton X-100. Block with 10% BSA. Incubate with primary antibodies (e.g., anti-GFP, anti-α-tubulin) overnight at 4°C. Label with secondary antibodies conjugated to STED-compatible dyes (e.g., Abberior STAR RED, Atto 594) for 1 hr at RT. Use phalloidin to label actin cortex.
- STED Imaging: Image using a STED microscope equipped with a 775 nm depletion laser. Acquire z-stacks encompassing the cell-cortex interface. Note the precise stage coordinates of grid squares and cells of interest.
- Post-fixation and Embedding for EM: Post-fix cells in situ with 2.5% glutaraldehyde in 0.1M cacodylate buffer for 1 hr. Perform osmication (1% osmium tetroxide) and en bloc staining (1% uranyl acetate). Dehydrate in an ethanol series and embed in EPON resin. Polymerize at 60°C for 48 hrs.
- Lamella Preparation: Using the finder grid coordinates, excise the region of interest. Trim the block and section using an ultramicrotome to obtain 70-100 nm ultrathin sections. Collect sections on TEM grids.
Protocol 2: Fiducial-Based Image Registration and Correlation Objective: Precisely align STED and TEM images of the same cellular region using fiduciary markers.
- Fiducial Application: After STED imaging but before osmication, apply a sparse layer of 100 nm gold fiducial beads (e.g., Aurion) with a known, irregular pattern onto the sample surface.
- Image Acquisition: Capture STED images containing both the cellular structures and the fiducial beads. After TEM sectioning, acquire low-magnification TEM overviews to locate the same fiducial beads and cellular region.
- Image Registration: Use correlation software (e.g., ec-CLEM plugin for Icy, Fiji). Manually select corresponding fiducial points (≥3) in both the STED and TEM image sets. Apply a landmark-based affine or projective transformation model.
- Validation & Analysis: Calculate the registration error (root-mean-square deviation of fiducial points). Overlay the transformed STED fluorescence channel onto the TEM micrograph. Quantify distances and co-localizations as per Table 2.
Visualizations
Diagram 1: The Correlative Validation Workflow (63 chars)
Diagram 2: Detailed CLEM Protocol Steps (94 chars)
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for STED-EM Correlation
| Item / Reagent | Function / Application | Key Consideration |
|---|---|---|
| Finder Grid Coverslips | Provides a coordinate system to relocate the same cell after embedding and sectioning. | Essential for reliable correlation. |
| Mild Crosslinker (0.1-0.5% Glutaraldehyde) | Preserves ultrastructure for EM while maintaining antigenicity for fluorescence. | Concentration is a critical balance. |
| STED-Optimized Secondary Antibodies (e.g., conjugated to Abberior STAR RED, ATTO 594) | Provides bright, photostable signal compatible with STED depletion lasers. | Red/far-red dyes minimize cellular autofluorescence. |
| 100 nm Gold Fiducial Beads | Unambiguous markers for precise software-based registration of STED and EM images. | Must be applied post-STED, pre-EM processing. |
| Tokuyasu Cryosectioning Reagents (Sucrose, Methylcellulose, Uranyl Acetate) | An alternative protocol offering superior antigen preservation for challenging antibodies. | Requires cryo-ultramicrotome. |
| Correlation Software (e.g., Icy ec-CLEM, Fiji/TrakEM2) | Aligns multi-modal image datasets using fiducial or landmark-based transformation. | Open-source solutions are available. |
| Low-Autofluorescence EPON Resin | Embedding medium for ultramicrotomy that minimizes background fluorescence retention. | Standard resins can quench fluorescence. |
Application Notes: The Super-Resolution Landscape
Understanding the dynamic interface between microtubule plus-ends and the cell cortex is critical for studying processes like cell division, polarization, and migration. Super-resolution microscopy is essential, as these interactions occur at a scale below the diffraction limit of light (~250 nm). This note compares key techniques within the context of studying these nanoscale events.
Table 1: Super-Resolution Modality Comparison for Microtubule-Cortex Imaging
| Modality | Effective Resolution (xy) | Key Advantage for Cortex Studies | Key Limitation | Optimal Use Case |
|---|---|---|---|---|
| STED | 30-80 nm | Targeted, on-demand super-resolution with high temporal control. Excellent for dense cortical arrays. | High illumination intensity. Potential photobleaching. | Quantifying microtubule end distance to cortex protein markers. |
| SIM | ~100 nm | Gentle on samples. Good for live-cell, multi-color imaging. | Moderate resolution gain. Reconstruction artifacts possible. | Observing relative dynamics of microtubules and cortical markers over time. |
| PALM/STORM | 20-50 nm | Highest achievable resolution. Molecular counting potential. | Very slow acquisition. Special buffers required. | Mapping ultrastructural architecture of cortical docking complexes in fixed cells. |
| Expansion Microscopy | ~70 nm (post-expansion) | Compatible with standard microscopes. Preserves ultrastructure. | Physical distortion concerns. No live-cell. | Correlative studies with EM or mapping many cortical components. |
Table 2: Decision Matrix for Tool Selection
| Primary Research Question | Recommended Primary Tool | Complementary Tool | Rationale |
|---|---|---|---|
| What is the nanoscale spatial organization of cortical anchoring proteins relative to microtubule ends? | PALM/STORM | Expansion Microscopy | Maximum resolution for static, molecular-scale mapping. |
| How do microtubule tip dynamics change upon cortical contact in live cells? | STED | TIRF (for pre-localization) | Balance of super-resolution speed and precision to track dynamic events. |
| What is the large-scale, 3D architecture of the microtubule cortex interaction zone? | SIM (3D) | Confocal Microscopy | Gentle, volumetric imaging of larger cellular regions. |
| Is protein 'X' part of the physical linker complex at the cortex? | STED with gSTED | FRET/FLIM | Use STED for colocalization analysis at sub-diffraction scale, then FRET for interaction proof. |
Detailed Experimental Protocols
Protocol 1: Sample Preparation for STED Imaging of Microtubule Plus-Ends
Objective: To label microtubules and cortical markers for dual-color STED imaging in fixed mammalian cells.
Materials: See "Research Reagent Solutions" table.
Procedure:
- Cell Culture & Plating: Plate cells (e.g., RPE-1, HeLa) on high-performance #1.5H coverslips. Culture to 60-70% confluency.
- Fixation & Permeabilization: Rinse cells briefly in pre-warmed PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9). Fix with 4% paraformaldehyde + 0.1% glutaraldehyde in PHEM for 10 min at 37°C. Critical: Low glutaraldehyde improves preservation but requires subsequent quenching with 0.1% NaBH4 for 7 min.
- Permeabilization & Blocking: Permeabilize with 0.5% Triton X-100 in PBS for 10 min. Block with 5% BSA + 0.1% Tween-20 in PBS (Blocking Buffer) for 1 hour.
- Immunostaining:
- Incubate with primary antibodies (e.g., anti-α-Tubulin, anti-CLIP170/EB1 for plus-ends, anti-Ezrin for cortex) diluted in Blocking Buffer overnight at 4°C.
- Wash 3x 5 min with PBS + 0.1% Tween-20 (PBST).
- Incubate with secondary antibodies conjugated to STED-compatible dyes (e.g., Abberior STAR 580 for microtubules, Abberior STAR 635P for cortex). Use dye separation of >80 nm for optimal STED. Incubate for 1 hour at RT, in darkness.
- Wash 3x 5 min with PBST.
- Mounting: Mount in ProLong Diamond Antifade mountant. Cure for 24 hours at RT before imaging.
Protocol 2: STED Image Acquisition for Cortical Microtubule Tips
Objective: To acquire super-resolved images of microtubule ends at the cell cortex.
Pre-imaging Setup:
- Align the STED depletion laser (775 nm typical) using a reference dye bead sample to ensure a perfect donut.
- On the sample, first locate the cell cortex using confocal mode with a 100x/1.4 NA oil objective.
Acquisition Parameters:
- Pinhole: 1 Airy Unit (confocal mode).
- Scanning Speed: 6-8 lines per second for optimal signal-to-noise.
- Pixel Size: 20 nm x 20 nm (Nyquist sampling for ~60 nm resolution).
- STED Laser Power: Titrate from 10-80% to find the minimum power that gives consistent resolution improvement without bleaching. Start at 30%.
- Sequential Acquisition: Acquire Channel 1 (microtubule, e.g., STAR580) with its corresponding STED laser, then Channel 2 (cortex, e.g., STAR635P) to avoid cross-talk.
- Frame Accumulation: 4-8 line accumulations to improve SNR.
Analysis Workflow:
- Deconvolution: Apply a GPU-accelerated deconvolution algorithm (e.g., Huygens) to raw STED data to further enhance resolution.
- Alignment: If using sequential acquisition, perform a sub-pixel channel alignment using multicolor bead images.
- Distance Measurement: Use line scan analysis orthogonal to the cortex to measure the distance between the peak of the microtubule end signal and the peak of the cortical marker signal.
Visualizing the Experimental Strategy
Title: STED Sample Prep and Imaging Workflow
Title: Microtubule-Cortex Interaction Logic
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Imaging Microtubule-Cortex Interactions
| Reagent Category | Specific Example(s) | Function in Experiment | Critical Notes |
|---|---|---|---|
| STED-Compatible Fluorophores | Abberior STAR 580, STAR 635P; ATTO 590, ATTO 647N | High-intensity, photostable dyes that can be efficiently depleted by the STED laser. | Ensure excitation/STED laser lines match your microscope. Separate emission spectra >80 nm. |
| Microtubule Plus-End Markers | Anti-EB1, Anti-CLIP170 antibodies; EB3-GFP live-cell construct | Specifically label the dynamic growing ends of microtubules to identify sites of cortical contact. | Fixed vs. live-cell requires different tools. Overexpression of fluorescent proteins can perturb dynamics. |
| Cortical Marker Antibodies | Anti-Ezrin/Radixin/Moesin (ERM), Anti-Anillin, Anti-Spectrin | Label the actin cortex and specific cortical proteins implicated in microtubule anchoring. | Validate specificity for your cell type. Choose markers relevant to your biological process (e.g., Anillin for cytokinesis). |
| Mounting Medium | ProLong Diamond, Abberior Mount Solid | Preserves fluorescence, reduces photobleaching under intense STED light, and maintains index matching. | For 3D STED, use a medium with refractive index matched to immersion oil (n=1.518). |
| Fiducial Markers | Tetraspeck beads, Gatta-PACT beads | For precise alignment of multiple color channels at the nanoscale. Essential for distance measurements. | Add a dilute solution to the sample before mounting. |
| Live-Cell Imaging Buffer | CO2-independent medium, HEPES-buffered, with antioxidants | Maintains cell health during time-lapse STED imaging, which can be phototoxic. | For prolonged imaging, use a system with environmental control (temp, CO2). |
Conclusion
STED super-resolution microscopy has emerged as a uniquely powerful tool for dissecting the nanoscale organization and dynamics of microtubule-cortex connections, a nexus critical for fundamental cell biology and implicated in disease. By providing a methodological roadmap—from foundational understanding through optimized application and rigorous validation—this guide empowers researchers to reliably capture these elusive interfaces. The comparative analysis underscores STED's particular strength in live-cell, multi-color imaging with direct, instrument-defined resolution. Future directions point toward integrating STED with traction force microscopy to correlate nanostructure with mechanics, and applying these protocols to drug discovery pipelines targeting the cytoskeleton in cancer and neurodegeneration. Mastering STED for this application will continue to yield transformative insights into cellular architecture and function.