Microtubule Lattice Dynamics: From Structural Heterogeneity to Clinical Translation
This article synthesizes recent breakthroughs in understanding microtubule lattice dynamics, moving beyond the traditional view of a static microtubule shaft.
Microtubule Lattice Dynamics: From Structural Heterogeneity to Clinical Translation
Abstract
This article synthesizes recent breakthroughs in understanding microtubule lattice dynamics, moving beyond the traditional view of a static microtubule shaft. We explore the foundational concepts of lattice heterogeneity, including seams and topological defects, and detail cutting-edge characterization methods like segmented subtomogram averaging and super-resolution microscopy. The review further examines how microtubule-associated proteins and drugs modulate lattice stability and turnover, and provides a critical comparison of in vitro versus cellular assays for drug development. Finally, we discuss the translational implications of lattice dynamics in neurodegenerative diseases and cancer therapy, offering a comprehensive resource for researchers and drug development professionals.
Beyond the Static Shaft: Unveiling Lattice Heterogeneity and Intrinsic Dynamics
Frequently Asked Questions (FAQs)
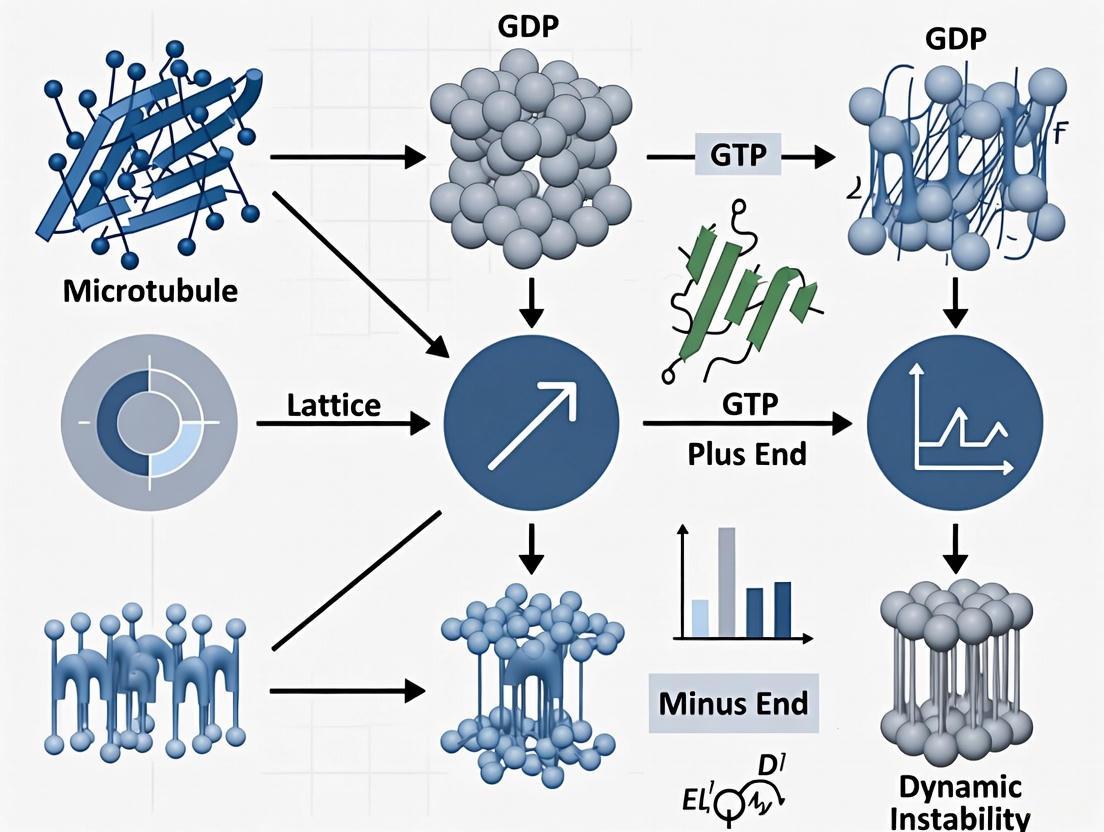
Q1: What are the two primary lattice types in microtubules, and which one is predominant in cytoplasmic microtubules? Microtubules can form two distinct lattice types: the A-lattice and the B-lattice. In the A-lattice, α-tubulin subunits are situated adjacent to β-tubulin subunits on neighboring protofilaments. In the B-lattice, the more common type, α-tubulin lies beside α-tubulin, and β-tubulin beside β-tubulin [1]. Research using cryo-electron tomography on cytoplasmic microtubules from mammalian cells has confirmed that the B-lattice is the predominant arrangement in vivo [1]. However, because microtubules with 13 protofilaments and a B-lattice cannot close into a perfect cylinder, they contain a single structural discontinuity called a "seam," where the tubulin subunits interact in an A-lattice configuration [2] [1].
Q2: My experiments on lattice spacing are yielding inconsistent results. What factors could be regulating this spacing? The microtubule lattice spacing, or dimer rise, is not static but a dynamic property regulated by several competing factors. Your results may vary due to:
- Nucleotide State: GTP-bound tubulin tends to form an expanded lattice (83.5 Ã…), while GDP-bound tubulin typically forms a compacted lattice (81.7 Ã…) [3].
- MAP Binding: Microtubule-associated proteins (MAPs) can directly influence spacing. For example, kinesin-1 acts as a lattice expander, while doublecortin (DCX) acts as a compactor [3].
- Pharmacological Agents: Drugs like paclitaxel stabilize and expand the microtubule lattice [3].
- Mechanical Forces: Bending or compressive forces can compress the lattice on the inner curve of a bend and expand it on the outer curve [3]. The observed lattice state in an experiment is often the result of the local balance between these expanding and compacting forces [3].
Q3: How does the neuronal protein tau affect microtubule lattice dynamics, beyond simple stabilization? Historically viewed as a passive stabilizer, tau is now known to actively modulate the microtubule lattice. Although it stabilizes microtubules against catastrophic fracture [4], tau surprisingly accelerates the exchange of tubulin dimers within the lattice itself [4]. This exchange occurs preferentially at topological defect sites. Tau achieves this by increasing lattice anisotropy—it stabilizes longitudinal tubulin-tubulin interactions while destabilizing lateral ones. This promotes the mobility and annihilation of lattice defects, effectively enabling the lattice to self-repair [4].
Q4: What is the functional significance of the microtubule seam? The seam is a unique long-range structural defect where the typical B-lattice transitions into an A-lattice. This break in helical symmetry means the microtubule is not a perfectly cylindrical structure and has two distinct faces [1]. This structural uniqueness has functional consequences; the seam can serve as a specific binding site for certain MAPs. For instance, the End-Binding protein EB1 has been shown to bind preferentially along the seam, which may help stabilize the microtubule structure in a specific orientation [1].
Troubleshooting Guides
Issue 1: Uninterpretable Kymographs from Microtubule Dynamics Assays
Problem: Kymographs show blurry or inconsistent microtubule trajectories, making it difficult to measure growth speeds or catastrophe frequencies.
Solution:
- Verify Preparation of Labelled Tubulin: Ensure the fluorescent tubulin is functional and free of aggregates. Use ultracentrifugation (e.g., at 100,000 x g for 10 minutes) to pellet aggregates immediately before use.
- Optimize Imaging Conditions: Reduce background noise by using TIRF microscopy. Shorten exposure times and increase laser intensity to "freeze" dynamic ends, but be mindful of photobleaching and phototoxicity.
- Confirm Assay Buffer Health: Ensure an adequate GTP-to-tubulin ratio (typically >1:1) and include an oxygen-scavenging system (e.g., glucose oxidase/catalase) and a triplet-state quencher (e.g., Trolox) to prolong filament and fluorophore longevity.
- Positive Control: Validate your entire workflow by reproducing a well-established result, such as the acceleration of microtubule growth by XMAP215/CLASP family proteins.
Issue 2: Failure to Recapitulate Lattice Spacing Phenomena from Literature
Problem: In vitro experiments with MAPs like DCX or drugs like paclitaxel do not produce the expected changes in microtubule lattice spacing or organization.
Solution:
- Re-check Protein Purification and Activity: Confirm that your recombinant MAP is purified, folded correctly, and is active in a standard microtubule co-sedimentation (pull-down) assay.
- Titrate Critical Components: The effect on lattice spacing is highly concentration-dependent. Systematically titrate both the MAP (e.g., DCX) and the drug (e.g., paclitaxel), as their influence is competitive [3]. A high concentration of a compactor like DCX can counteract the expanding effect of paclitaxel [3].
- Control Nucleotide State: Use non-hydrolysable GTP analogues (e.g., GMPCPP) to create purely expanded lattices or GDP with specific salts to create compacted lattices as baseline controls for your assays [3].
- Use a Sensitive Readout: If direct cryo-EM is not feasible, employ an indirect functional assay. For example, monitor the re-localization of a GFP-tagged compactor like DCX in cells upon paclitaxel treatment, as its binding is lattice-spacing-dependent [3].
Issue 3: High Background in Tubulin Lattice Incorporation Assays
Problem: Excessive fluorescent background obscures the specific signal from tubulin dimers incorporated into the microtubule lattice.
Solution:
- Refine Wash Steps: After incubating with fluorescent tubulin, implement rigorous wash procedures. Use multiple flows of warm assay buffer (at least 5-10 chamber volumes) to remove unincorporated tubulin dimers thoroughly.
- Optimize Concentrations: Reduce the concentration of free fluorescent tubulin during the incorporation phase. The goal is to favor incorporation at lattice defect sites over spontaneous nucleation in solution. Test concentrations in the range of 5-10 µM [4].
- Include a Capping Agent: To prevent incorporation at the microtubule ends from dominating the signal, cap the ends with stable, unlabeled GMPCPP seeds or a plus-end-binding protein that inhibits growth [4].
- Confirm Microtubule Stabilization: Ensure the "stable" microtubule seeds are genuinely inert. Polymerize seeds with GMPCPP and verify their stability by imaging over time in the absence of free tubulin.
Quantitative Data Tables
Table 1: Experimentally Observed Microtubule Lattice Spacing
This table summarizes key quantitative measurements of microtubule lattice spacing under different conditions, crucial for interpreting structural data.
| Nucleotide State / Condition | Lattice Spacing (Å, mean ± SEM) | Lattice Conformation | Primary Experimental Method | Key Reference Context |
|---|---|---|---|---|
| GTP-like State (GMPCPP) | 83.5 ± 0.2 | Expanded | Cryo-EM | [3] |
| GDP State (in vitro) | 81.7 ± 0.1 | Compacted | Cryo-EM | [3] |
| + Microtubule Expander (e.g., Kinesin-1) | ~83.5 | Expanded | Light Microscopy / Cryo-EM | [3] |
| + Microtubule Compactor (e.g., DCX) | ~81.7 | Compacted | Cryo-EM / X-ray Diffraction | [3] |
Table 2: Quantifying the Impact of Tau on Microtubule Lattice Dynamics
This table presents quantitative data on how the MAP tau influences tubulin exchange and mechanical integrity of the microtubule lattice.
| Experimental Parameter | Control (0 nM Tau) | With 20 nM Tau | Change | Experimental Context |
|---|---|---|---|---|
| Median Tubulin Incorporation Length (after 15 min) | 0.7 µm | 1.2 µm | +71% | In vitro incorporation assay [4] |
| Spatial Frequency of Incorporation (median distance between events) | 12.4 µm | 6.6 µm | -47% | In vitro incorporation assay [4] |
| Overall Tubulin Incorporation (after 15 min) | 1x (baseline) | 4x | +300% | In vitro incorporation assay [4] |
| Lattice Fluorescence Loss at Incorporation Sites (after 30 min) | ~7% | ~15% | Increased loss | Indicates tubulin exchange, not just addition [4] |
| Microtubule Fracture Rate (in absence of free tubulin) | 1x (baseline) | Slower | Decreased | Enhanced mechanical stability [4] |
Experimental Protocols
Protocol 1: Determining Microtubule Lattice Type via Cryo-Electron Tomography
Objective: To directly visualize the lattice structure and identify the seam in cellular cytoplasmic microtubules.
Background: This protocol is adapted from studies that resolved the B-lattice structure of microtubules in mammalian cells [1]. It involves decorating microtubules with a motor protein to reveal the underlying tubulin dimer arrangement.
Materials:
- Cultured cells (e.g., 3T3 fibroblasts) grown on carbon-coated electron microscopy grids.
- Lysis/Permeabilization Buffer: 60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgClâ‚‚, 0.1% Triton X-100, pH 6.9.
- Purified monomeric motor domain of Eg5 (kinesin-5).
- Liquid ethane for plunge-freezing.
- Cryo-electron microscope equipped with a tilting holder.
Method:
- Cell Lysis and Decoration: Quickly perfuse the lysis buffer containing ~0.1 mg/mL of the Eg5 motor domain over the cells on the grid for 20-30 seconds. This permeabilizes the cells while preserving microtubules and allows motor proteins to bind to them [1].
- Rapid Freezing: Blot the grid to remove excess liquid and immediately plunge-freeze it in liquid ethane to preserve the structure in a vitreous ice state.
- Tomographic Data Collection: Transfer the grid to the cryo-electron microscope. Collect a tilt-series of images (e.g., from -60° to +60° at 2° intervals) of cellular regions where microtubules span holes in the carbon film.
- Image Reconstruction and Analysis: Use software (e.g., IMOD) to align the tilt-series and reconstruct a 3D tomogram. Inspect the tomogram for the helical pattern of the bound motor heads. A single, paraxial seam and a pattern consistent with a B-lattice (where motors follow a left-handed, 1.5-start helix) confirm the predominant lattice structure [1].
Protocol 2: In Vitro Assay for Microtubule Lattice Spacing Competition
Objective: To investigate the competitive regulation of microtubule lattice spacing by a compactor (DCX) and an expander (paclitaxel).
Background: This assay, based on recent research, uses microtubule buckling as a readout for lattice spacing changes observable by light microscopy [3]. An expanded lattice will buckle more under constrained conditions.
Materials:
- Purified tubulin.
- Microtubule compactor protein (e.g., Doublecortin/DCX).
- Microtubule expander drug (e.g., Paclitaxel).
- Non-hydrolysable GTP analogue (GMPCPP).
- Flow chamber for microscopy.
- TIRF microscope.
Method:
- Form Double-Capped Microtubules: Polymerize microtubules with a central GDP-lattice segment capped at both ends by stable GMPCPP-lattice sections [3].
- Immobilize and Induce Compression: Anchor these microtubules to a coverslip surface. The inherent length difference between the GDP and GMPCPP lattices upon introduction to assay buffer will generate axial compression on the central GDP segment.
- Apply Test Conditions: Introduce the compactor (DCX) and/or expander (paclitaxel) at varying concentrations.
- Image and Quantify Buckling: Use TIRF microscopy to record microtubule behavior. The frequency and amplitude of buckling events in the central segment serve as a proxy for lattice expansion. For example, high paclitaxel concentrations should induce more buckling (expansion), which can be suppressed by sufficiently high concentrations of DCX (compaction) [3].
Research Reagent Solutions
Table 3: Essential Reagents for Microtubule Lattice Characterization
| Reagent Name | Function / Description | Key Application in Lattice Research |
|---|---|---|
| GMPCPP (Guanylyl-(α,β)-methylene-diphosphonate) | A non-hydrolysable GTP analog that stabilizes microtubules in a GTP-like, expanded lattice state [3] [4]. | Creating stable seeds for polymerization; studying expanded lattice structure and dynamics. |
| Paclitaxel (Taxol) | A small molecule drug that binds and stabilizes microtubules, promoting an expanded lattice conformation [3]. | A tool to experimentally induce and study expanded lattice states; used in competition experiments with compactors. |
| Doublecortin (DCX) | A neuronal Microtubule-Associated Protein (MAP) that functions as a lattice compactor, stabilizing a compacted state [3]. | A tool to experimentally induce and study compacted lattice states; used to investigate competition for lattice spacing control. |
| Tau Protein (e.g., 2N4R isoform) | A neuronal MAP that stabilizes microtubules but accelerates tubulin exchange within the lattice, facilitating defect repair [4]. | Studying lattice dynamics, self-repair mechanisms, and the role of MAPs beyond simple stabilization. |
| Kinesin-1 Motor Domain | A molecular motor that binds processively to microtubules and can act as a lattice expander [3]. | Probing lattice state (expanded vs. compacted); studying the interplay between motor proteins and lattice structure. |
| Monomeric Eg5 Motor Domain | A kinesin motor domain that binds densely to the microtubule lattice without processive movement, decorating the underlying tubulin arrangement [1]. | A marker for visualizing the helical tubulin lattice and identifying the seam in structural studies (e.g., cryo-ET). |
Signaling Pathways & Workflow Diagrams
Tau-Mediated Lattice Repair
Lattice Spacing Competition
The Role of Seams and Multi-Seam Structures in Lattice Integrity
Troubleshooting Guide & FAQ
Frequently Asked Questions
Q1: What is a microtubule lattice seam, and why is it important for researchers to identify? A lattice seam is a structural discontinuity in the microtubule wall where protofilaments associate via heterotypic (α-β) lateral contacts, unlike the homotypic (α-α or β-β) contacts found in the rest of the B-lattice [5]. Accurately determining its location is critical because it breaks the helical symmetry, and misidentification can lead to incorrect modeling of key functional regions, such as the nucleotide state at the E-site in β-tubulin and the geometry of lateral contacts [5]. For dynamic instability studies, the seam can act as a trigger point for catastrophe [6].
Q2: Our cryo-EM processing of microtubules is failing to reach a consensus on seam location. What could be the cause and solution? This is common when the marker protein used for registration is relatively small or its decoration is sparse [5]. Traditional projection-matching methods can fail under these conditions. A solution is to implement a specialized seam-search protocol that leverages the intrinsic ~80 Å tubulin dimer repeat signal in the raw images, which can help determine the αβ-tubulin register even without a large marker [5].
Q3: We are observing unusually high shrinkage rates and catastrophe frequency in our microtubule assays. Could the lattice structure be a factor? Yes. Experimental evidence shows that microtubules with extra A-lattice seams are significantly destabilized. For example, GMPCPP-stabilized microtubules with enriched A-lattice content shrink at a median rate over 20 times faster than their B-lattice counterparts [6]. Introducing multiple seams creates pre-existing pathways that accelerate damage propagation and destabilize the lattice [7].
Q4: How can we experimentally produce microtubules with defined seam numbers for comparative studies? While spontaneous in vitro assembly rarely produces such microtubules, you can use the S. pombe EB1 protein Mal3. Adding a high concentration of Mal3 during nucleation can drive the assembly of A-lattice-enriched 13-protofilament microtubules. It is crucial to remove Mal3 after assembly before stability assays, as lattice-bound Mal3 can itself stabilize microtubules [6].
Troubleshooting Common Experimental Issues
| Problem | Potential Cause | Recommended Solution |
|---|---|---|
| Unstable 3D Reconstructions | Incorrect initial helical parameters or seam location [5] | Perform multi-reference alignment against models with different protofilament numbers (e.g., 12-15 PFs) to determine initial parameters [5]. |
| Inability to Distinguish α/β-tubulin | Lack of a clear structural marker in cryo-EM images [5] | Employ a dedicated seam-search strategy that utilizes the tubulin dimer repeat signal, even with small marker proteins like EB's CH domain [5]. |
| High Catastrophe Frequency | Underlying lattice defects or multiple seams in seeds [6] | Verify the seam structure of nucleating seeds. Use B-lattice seeds with a single seam for more stable growth [6]. |
| Excessive Microtubule Shrinkage | A-lattice enriched structures destabilizing the lattice [6] | Control for seam content during polymerization. Assess shrinkage rates against a known B-lattice single-seam control [6]. |
| Microtubule Fracture/Damage | Low ratio of longitudinal to lateral binding energies; multiple seams [7] | Ensure proper buffer conditions. Be aware that simulations suggest the longitudinal/lateral binding energy ratio is bounded near 1.5 for stability [7]. |
Table 1: Impact of A-Lattice Seam Content on Microtubule Stability
This table summarizes key quantitative findings on how seam content influences microtubule dynamic instability, based on experimental data [6].
| Microtubule Type / Seed | A-Lattice Content | Median Shrinkage Rate (nm/s) | Catastrophe Frequency | Key Experimental Condition |
|---|---|---|---|---|
| B-lattice Single-seam | ~1 seam | 2.5 | Baseline (reference) | GMPCPP, pig brain tubulin [6] |
| A-lattice Enriched (Mal3-N143) | ~50% (avg.) | 22.6 | Increased | GMPCPP, assembled with monomeric Mal3-CH [6] |
| A-lattice Enriched (Mal3FL) | ~50% (avg.) | 58.8 | Increased | GMPCPP, assembled with dimeric full-length Mal3 [6] |
| Dynamic MTs (A-lattice seeds) | Multiple seams | Growth rate similar to B-lattice | Increased at both ends | Nucleated from A-lattice enriched seeds [6] |
Table 2: Key Measurements for Structural Analysis
This table outlines critical measurements for characterizing microtubule lattice structure, drawing parallels from precise seam inspection methodologies [8].
| Measurement | Description | Significance in Lattice Integrity |
|---|---|---|
| Protofilament (PF) Number | The number of protofilaments forming the microtubule wall [5] | Defines the basic lattice type and curvature; influences mechanical strength [5]. |
| Seam Location | The specific protofilament interface with heterotypic (α-β) contacts [5] | Critical for correct helical symmetry imposition in 3D reconstruction [5]. |
| Lateral Contact Type | Geometry of interactions (B-lattice: α-α/β-β; Seam: α-β) [5] [6] | A-lattice contacts at seams may have different stability compared to B-lattice [6]. |
| Nucleotide State (E-site) | GTP vs. GDP in β-tubulin, visualized in high-res maps [5] | Directly reports on the stability of the microtubule; GDP-core is unstable [5] [6]. |
| Body Wall Thickness | The physical thickness of the polymer wall. | An analog to fundamental structural integrity measurements [8]. |
Detailed Experimental Protocols
Protocol 1: Kinesin Surface Clamp-Release Assay for Microtubule Stability
Purpose: To measure the shrinkage rates and stability of microtubules with different seam contents after release from a stabilizing rigor kinesin coat [6].
Materials:
- GMPCPP-stabilized microtubules (B-lattice or A-lattice enriched)
- Kinesin-1 motor proteins (non-processive, single-head constructs are suitable)
- Flow chamber with coverslip
- BRB80 or similar microtubule-stabilizing buffer
- ATP-containing buffer (e.g., BRB80 + 1mM ATP)
- TIRF or epifluorescence microscope
Method:
- Surface Preparation: Coat the flow chamber with a rigor kinesin solution (lacking ATP) to create a stable, non-motile kinesin layer.
- Microtubule Binding: Introduce fluorescently-labeled GMPCPP microtubules into the chamber. Allow them to bind to the kinesin-coated surface in ATP-free conditions. This "clamps" and stabilizes the MTs.
- Washing: Flush the chamber extensively with ATP-free buffer to remove any unbound microtubules and, critically, any residual Mal3 if A-lattice-enriched MTs were used [6].
- Release and Imaging: Rapidly flush the chamber with buffer containing 1mM ATP. This activates the kinesin motors, causing them to walk and releasing the microtubules from the stable clamp.
- Data Acquisition: Immediately acquire time-lapse images to track the lengths of the freed microtubules over time as they depolymerize.
Data Analysis:
- Measure microtubule length frame-by-frame.
- Plot length vs. time to calculate shrinkage rates (nm/s) for both plus and minus ends.
- Compare median shrinkage rates between B-lattice single-seam and A-lattice-enriched microtubules.
Protocol 2: Cryo-EM Seam Location and Reconstruction
Purpose: To accurately determine the αβ-tubulin register and seam location for each microtubule segment during single-particle cryo-EM processing [5].
Materials:
- Cryo-EM grid of microtubule sample (e.g., decorated with kinesin or EB)
- Processing software (e.g., FREALIGN, EMAN1/2, RELION)
- Access to a high-performance computing cluster
Method:
- Data Collection & Pre-processing: Collect movie-mode data on a direct electron detector. Perform drift and motion correction. Estimate the contrast transfer function (CTF) for each micrograph [5].
- Particle Picking: Manually or semi-automatically select microtubule segments from the micrographs. Extract these as overlapping boxes, with a non-overlapping region set to ~80 Ã… (the tubulin dimer repeat) [5].
- Initial Alignment & PF Number Determination: Compare raw particles to projections of low-pass-filtered reference models with different protofilament numbers (e.g., 12, 13, 14, 15 PFs). Use multi-reference alignment to determine the initial global orientation and correct PF number for each particle [5].
- Seam-Search Strategy: Refine alignment parameters using a local refinement algorithm. To determine the seam location, use specialized strategies such as:
- Projection Matching: Compare raw segments to projections of reference models with all possible seam locations and identify the one with the highest cross-correlation.
- Dimer Repeat Signal: Utilize the intrinsic power of the ~80 Ã… tubulin dimer repeat in the raw images to inform the seam search, which is particularly useful when marker protein signal is weak [5].
- 3D Reconstruction: Merge particles with the same PF number and refined seam location. Reconstruct the 3D density map using either pseudo-helical symmetry (applying symmetry operations but skipping the seam) or no symmetry (C1) [5].
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function/Explanation | Application in Seam Research |
|---|---|---|
| Mal3 (EB1 Homolog) | S. pombe end-binding protein. Used as a tool to nucleate microtubules with enriched A-lattice content when added at high concentrations [6]. | Generating defined A-lattice seam structures for stability assays. |
| GMPCPP Tubulin | Assembled from tubulin and a non-hydrolysable GTP analog. Forms microtubules that are structural analogues of the stabilizing GTP cap [6]. | Testing the intrinsic stability of different lattice architectures without complication from dynamic instability. |
| Rigor Kinesin | A kinesin motor mutant or state (e.g., lacking ATP) that binds tightly to microtubules without moving. | Stabilizing microtubules in surface assays for buffer exchange and controlled release [6]. |
| Direct Electron Detector | A camera for cryo-EM that allows for movie-mode data collection, enabling motion correction and high-resolution reconstruction [5]. | Essential for achieving the resolution needed to distinguish α/β-tubulin and identify seam location. |
| Iterative Helical Real Space Reconstruction (IHRSR) | A cryo-EM image processing algorithm modified for microtubules that iteratively refines helical parameters [5]. | Determining the 3D structure of microtubules, including those with a lattice seam. |
| A-770041 | A-770041, CAS:1140478-96-1, MF:C34H39N9O3, MW:621.7 g/mol | Chemical Reagent |
| RGD peptide (GRGDNP) (TFA) | RGD peptide (GRGDNP) (TFA), MF:C25H39F3N10O12, MW:728.6 g/mol | Chemical Reagent |
Experimental Workflow & Pathway Diagrams
Topological Defects as Hotspots for Tubulin Exchange and Lattice Remodeling
Frequently Asked Questions (FAQs)
1. What are topological defects in the microtubule lattice, and why are they important? Topological defects are structural imperfections in the otherwise ordered microtubule lattice, such as sites where protofilament number changes or at seam dislocations [4]. These defects are crucial because they act as hotspots for biological activity, serving as preferred locations for tubulin dimer exchange and incorporation [4] [9]. They weaken the local lattice structure by disrupting tubulin-tubulin interactions, which in turn drives lattice dynamics, self-repair, and remodeling [4] [10].
2. How does the protein tau influence microtubule lattice dynamics? Contrary to its traditional role as a passive stabilizer, recent research shows that tau actively accelerates tubulin exchange within the microtubule lattice despite having no enzymatic activity [4] [11] [12]. Tau binds to the microtubule shaft and modulates lattice dynamics by stabilizing longitudinal tubulin-tubulin interactions while simultaneously destabilizing lateral ones. This increases lattice anisotropy and promotes the mobility and annihilation of topological defects, effectively facilitating lattice remodeling and self-repair [4].
3. My stabilized microtubules are fracturing during experiments. What could be causing this? Lattice fracture in end-stabilized microtubules is an intrinsic property of the GDP-lattice and occurs even in the absence of external forces or free tubulin [10]. Fracture typically initiates at pre-existing topological defects or monomer vacancies and propagates through the lattice [10]. The time to fracture is usually between 10 and 20 minutes, with the damaged region spanning about 1 µm along the microtubule axis before breaking [10]. Incorporating the protein tau has been shown to slow down the fracture process, as it promotes defect repair [4].
4. What is the relationship between GTP hydrolysis and lattice dynamics? The prevailing "interface-acting" (trans) model indicates that the nucleotide at the interface between tubulin dimers—not the nucleotide bound to an individual tubulin—controls the strength of tubulin-tubulin interactions [13]. GTP hydrolysis after incorporation into the lattice weakens these interactions, creating a strained GDP-lattice that is primed for dynamics. This strained state makes defect sites particularly susceptible to tubulin loss and exchange [14] [13].
Troubleshooting Guides
Problem: Inconsistent Tubulin Incorporation in Lattice Incorporation Assays
Potential Cause and Solution:
- Cause: Variations in the density and distribution of inherent topological defects in your microtubule preparations can lead to inconsistent incorporation patterns [4] [10].
- Solution:
- Standardize Microtubule Seeds: Use GMPCPP-stabilized seeds to ensure a uniform nucleation template [4].
- Include a Regulator Protein: Incorporate a low concentration (e.g., 20 nM) of tau protein in your incorporation step. Tau homogenizes incorporation by preferentially targeting and enhancing exchange at defect sites [4].
- Control Incubation Time: Limit the incubation time with labelled tubulin (e.g., 15 minutes) to prevent saturation of incorporation sites, which can make quantification difficult [4].
Problem: Excessive Microtubule Fracture During Observation
Potential Cause and Solution:
- Cause: The intrinsic instability of the GDP-tubulin lattice and the presence of multiple seam structures accelerate damage propagation [10].
- Solution:
- Reinforce the Lattice: Include tau protein (20 nM) in your imaging buffer. Tau slows down fracture by promoting defect repair, even in the absence of free tubulin [4].
- Minimize Photodamage: Ensure your imaging parameters (e.g., laser power, exposure time) are optimized to avoid exacerbating lattice damage.
- Quantify Fracture Kinetics: Use your experimental setup to measure the time to fracture. Compare your results to established baselines (typically 10-20 minutes) to diagnose if fracture is occurring faster than expected due to experimental conditions [10].
Table 1: Key Parameters from Microtubule Lattice Fracture Studies
| Parameter | Experimental Value | Context / Condition |
|---|---|---|
| Time to Fracture | 10 - 20 minutes | For end-stabilized GDP-microtubules in the absence of free tubulin [10]. |
| Fracture Region Size | ~1 µm | Average length of the damaged lattice region before breakage [10]. |
| Lattice Anisotropy (A) | ~1.5 | Ratio of longitudinal to lateral binding energy ((A = \Delta G{\text{long}} / \Delta G{\text{lat}})) derived from fracture simulations [10]. |
Table 2: Effect of Tau on Lattice Incorporation Metrics
| Metric | Condition (15 min incubation) | Value |
|---|---|---|
| Median Incorporation Length | 0 nM Tau | 0.7 µm [4] |
| 20 nM Tau | 1.2 µm [4] | |
| Median Distance Between Incorporations | 0 nM Tau | 12.4 µm [4] |
| 20 nM Tau | 6.6 µm [4] | |
| Tubulin Loss from Original Lattice | 0 nM & 20 nM Tau | 7% - 15% reduction in fluorescence intensity [4] |
Experimental Protocols
Protocol 1: Studying Tau-Stimulated Tubulin Incorporation at Defect Sites
This protocol details how to visualize and quantify how tau promotes tubulin exchange at topological defects in the microtubule lattice [4].
Workflow Diagram: Tau-Stimulated Tubulin Incorporation Assay
Materials:
- Microtubule Seeds: GMPCPP-stabilized, surface-attached [4].
- Tubulin: Green-labeled GTP-tubulin (for initial growth), red-labeled GTP-tubulin (for incorporation assay), and unlabeled GMPCPP-tubulin (for capping) [4].
- Regulator Protein: Human 2N4R tau [4].
- Imaging Buffer: Tubulin-free buffer for final imaging [4].
Step-by-Step Procedure:
- Grow Microtubules: Dynamically grow microtubules from the surface-attached seeds using green-labelled GTP-tubulin [4].
- Cap Microtubule Tips: Inhibit further tip dynamics by capping the microtubules with slowly hydrolysable GMPCPP-tubulin. This isolates lattice dynamics from tip dynamics [4].
- Incorporation Step: Incubate the capped microtubules with a solution containing red-labelled GTP-tubulin (e.g., 8 µM) and your chosen concentration of tau (e.g., 0 nM for control, 0.5 nM, or 20 nM) for a defined period (e.g., 15 or 30 minutes) [4].
- Wash and Image: Wash out free tubulin to reduce background fluorescence. Image the microtubules to visualize the stretches of incorporated red tubulin. To correlate incorporation sites with weakness, continue imaging in tubulin-free buffer until microtubule fracture occurs [4].
Key Observations:
- In the presence of 20 nM tau, you should observe longer and more frequent stretches of incorporated tubulin [4].
- Approximately 40% of fracture events will occur at these incorporation stretches, identifying them as pre-existing defect sites [4].
- A slight decrease (7-15%) in the original (green) lattice fluorescence at incorporation sites indicates exchange (replacement) rather than simple addition of tubulin [4].
Protocol 2: Quantifying Lattice Anisotropy from Fracture Kinetics
This protocol uses a kinetic Monte Carlo model to deduce fundamental lattice energy parameters by simulating microtubule fracture [10].
Workflow Diagram: Lattice Anisotropy Simulation
Model Setup:
- Lattice Representation: Model the microtubule as a 2D lattice at the monomer scale, representing a 13-protofilament B-lattice with a possible A-lattice seam [10].
- Energy Parameters: Define the binding energies for longitudinal ((\Delta G{\text{long}})) and lateral ((\Delta G{\text{lat}})) interactions. The total binding energy for a fully surrounded dimer is (\Delta G{\text{b}} = 2\Delta G{\text{long}} + 2\Delta G_{\text{lat}}) [10].
- Anisotropy Definition: The key parameter, lattice anisotropy, is defined as (A = \Delta G{\text{long}} / \Delta G{\text{lat}}) [10].
- Detachment Kinetics: The detachment rate (k{mn}) of a dimer with (m) longitudinal and (n) lateral neighbors is given by: (k{mn} = \frac{1}{\tau} e^{\beta(m\Delta G{\text{long}} + \frac{n}{2}\Delta G{\text{lat}} - \frac{\Delta G_{\text{b}}}{2})}) where (\tau) is the off-rate for a corner dimer, and (\beta = 1/kT) [10].
Simulation and Analysis:
- Introduce Defect: Start with a 10 µm long, end-stabilized microtubule and introduce an initial monomer vacancy at a specific lattice position [10].
- Run Simulation: Simulate the progression of tubulin loss from this initial defect. The damage will typically propagate longitudinally faster than laterally if (A > 1) [10].
- Compare to Experiment: Adjust the anisotropy parameter (A) in your model so that the simulated time to fracture and damage propagation pattern match experimental observations (e.g., fracture in 10-20 minutes). Recent studies suggest (A) is bounded at approximately 1.5, indicating a less anisotropic lattice than previously thought [10].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Studying Microtubule Lattice Dynamics
| Reagent / Material | Function in Research | Key Details / Examples |
|---|---|---|
| GMPCPP-tubulin | A slowly hydrolysable GTP analogue. Used to create stable microtubule "seeds" for nucleation and to "cap" microtubule ends, allowing the study of pure lattice dynamics isolated from tip dynamics [4]. | Non-hydrolysable nucleotide analogue [4]. |
| Human 2N4R Tau | A specific isoform of the microtubule-associated protein tau. Used to investigate how MAPs modulate lattice dynamics, particularly in accelerating tubulin exchange and remodeling defects [4]. | Binds laterally along the microtubule lattice [4]. |
| Kinetic Monte Carlo Model | A computational model used to simulate the stochastic process of tubulin loss and incorporation. Essential for deducing energy parameters (like anisotropy) from experimental data on lattice fracture and exchange [10]. | Represents the MT as a 2D lattice; uses Arrhenius kinetics for detachment rates [10]. |
| End-Stabilized Microtubule Construct | A microtubule whose dynamic ends are chemically or structurally blocked. This is the fundamental experimental setup for studying intrinsic lattice dynamics without the confounding effects of tip growth or shrinkage [4] [10]. | Created by capping with GMPCPP-tubulin [4]. |
| Interference Reflection Microscopy | An imaging technique that allows visualization of microtubules without fluorescent labels. Used to control for potential artifacts introduced by fluorescent tags in lattice dynamics experiments [4]. | Validates findings from fluorescence microscopy [4]. |
| Teicoplanin A2-4 | Teicoplanin A2-4, MF:C89H99Cl2N9O33, MW:1893.7 g/mol | Chemical Reagent |
| FT-1518 | FT-1518, MF:C20H26N8O, MW:394.5 g/mol | Chemical Reagent |
FAQs: Core Concepts and Mechanisms
Q1: What is the fundamental difference between the cis-acting (self-acting) and trans-acting (interface-acting) models of nucleotide function in microtubules?
The fundamental difference lies in which nucleotide controls the strength of the tubulin:tubulin interaction.
- Cis-acting (Self-acting) Model: The nucleotide (GTP or GDP) bound to a specific tubulin dimer dictates how strongly that same dimer interacts with the microtubule lattice. It was historically thought that GTP induces a straight conformation for strong binding, while GDP induces a curved conformation for weak binding [15] [13].
- Trans-acting (Interface-acting) Model: The nucleotide at the interface between two tubulin dimers controls the binding strength. The nucleotide bound to the lower dimer in the lattice influences how tightly the upper dimer above it binds [16] [15]. This model is supported by structural data showing that both GTP- and GDP-tubulin adopt a curved conformation when unpolymerized [15] [13].
Q2: How does the nucleotide state (GDP vs. GTP) ultimately lead to microtubule catastrophe?
Catastrophe is initiated by the loss of the protective "GTP cap." Within the microtubule lattice, GTP is hydrolyzed to GDP. GDP-bound tubulin forms a labile lattice with weaker lateral contacts. When the GTP cap is lost and GDP-tubulin becomes exposed at the microtubule end, the weakened interactions can no longer sustain growth, prompting a rapid transition to shrinkage [16].
Q3: Why does GDP-tubulin "poison" microtubule growth when added to the solution, and which end is more affected?
GDP-tubulin has a lower affinity for the microtubule end than GTP-tubulin. When GDP-tubulin is incorporated into a growing end, it creates a weak point that destabilizes the structure. This "poisoning" effect is disproportionately stronger at the plus-end than at the minus-end. This end-specificity provides key experimental evidence for the interface-acting (trans) mechanism, as the plus-end has a distinct nucleotide configuration that is more susceptible to disruption by GDP [15] [13].
Q4: What is the role of nucleotide exchange in microtubule dynamics?
GDP-to-GTP exchange on terminal GDP-bound tubulin subunits at the microtubule end can mitigate the catastrophe-promoting effects of GDP exposure. By allowing a "weak" GDP-bound terminal subunit to be converted back to a "strong" GTP-bound state, nucleotide exchange helps maintain the integrity of the GTP cap and reduces the frequency of catastrophe [16] [15].
Troubleshooting Guides
Issue 1: High Catastrophe Frequency In Vitro
Problem: Your reconstituted microtubules undergo catastrophe too frequently, making it difficult to observe sustained growth or gather reliable dynamic instability parameters.
Potential Causes and Solutions:
- Cause: Low effective concentration of active GTP-tubulin.
- Solution: Increase the total tubulin concentration. Confirm the ratio of GTP to tubulin is sufficient (typically a molar excess of GTP) and prepare fresh tubulin in GTP-containing buffer just before the experiment.
- Cause: Excessive incorporation of GDP-tubulin.
- Solution: Ensure your tubulin preparation is of high quality and free of contaminating GDP. Use a rapid purification protocol or purchase commercial high-purity tubulin. Experimentally, using a slowly-hydrolyzable GTP analog (e.g., GMPCPP) can suppress catastrophe entirely for diagnostic purposes [15] [13].
- Cause: Unfavorable buffer conditions.
- Solution: Optimize buffer composition, specifically Mg²⺠concentration, pH, and the presence of glycerol or other stabilizing agents.
Issue 2: No Observable Differential Effect of GDP on Plus vs. Minus Ends
Problem: When performing mixed-nucleotide assays with GDP-tubulin, you do not observe the predicted disproportionate slowing of plus-end growth compared to the minus-end.
Potential Causes and Solutions:
- Cause: Inadequate suppression of GTP hydrolysis and catastrophe.
- Cause: Difficulty in reliably distinguishing and tracking the two ends.
- Solution: Use fiduciary markers (e.g., GMPCPP seeds) with distinct geometries to unambiguously identify plus and minus ends during data analysis.
Table 1: Key Parameters from Computational Models of Microtubule Dynamics
| Parameter | Cis-acting (Self-acting) Model | Trans-acting (Interface-acting) Model | Notes |
|---|---|---|---|
| GTPase Rate Requirement | Faster GTPase rate to achieve benchmark catastrophe frequency [16] | Slower GTPase rate to achieve the same catastrophe frequency [16] | The trans-acting model requires slower hydrolysis to explain observed dynamics. |
| GDP Effect on Plus-End | Moderate, proportional decrease in growth rate [15] [13] | Strong, disproportionate decrease in growth rate ("poisoning") [15] [13] | A key testable difference between the models. |
| GDP Effect on Minus-End | Proportional decrease in growth rate [15] [13] | Proportional decrease in growth rate (identical to cis-model) [15] [13] | The effect is identical for both models at the minus-end. |
| Impact of Nucleotide Exchange | Not a primary feature in early models | Reduces catastrophe frequency by mitigating GDP-poisoning [16] [15] | Incorporated into newer models to match experimental data. |
Table 2: Essential Research Reagents and Their Functions
| Reagent | Function in Experimentation |
|---|---|
| GMPCPP (slowly-hydrolyzable GTP analog) | Generates stable, non-dynamic microtubule seeds; used in mixed-nucleotide assays to suppress catastrophe and isolate the effects of GDP on pure elongation [15] [13]. |
| Mutant αβ-tubulin (with altered nucleotide affinity) | Used to selectively perturb the rate of GDP-to-GTP exchange on the microtubule end, testing the hypothesis that exchange influences catastrophe frequency [16]. |
| GDP-tubulin | Used in "mixed-nucleotide" assays to directly test the poisoning effect of incorporated GDP subunits on microtubule growth and to discriminate between cis and trans mechanisms [15] [13]. |
Experimental Protocols
Protocol 1: Mixed Nucleotide Assay to Distinguish Nucleotide Action Mechanisms
Objective: To experimentally determine whether nucleotide action follows a self-acting or interface-acting mechanism by measuring the differential effect of GDP-tubulin on plus-end vs. minus-end growth rates.
Background: This protocol tests a key prediction: interface-acting mechanisms cause GDP-tubulin to disproportionately inhibit plus-end growth compared to minus-end growth, while self-acting mechanisms inhibit both ends equally [15] [13].
Materials:
- Purified tubulin
- GMPCPP
- GDP
- GTP
- BRB80 buffer (80 mM PIPES, 1 mM MgClâ‚‚, 1 mM EGTA, pH 6.8)
- Flow chamber slides
- TIRF or fluorescence microscope
Method:
- Stabilized Seed Preparation: Polymerize tubulin in the presence of GMPCPP to form stabilized microtubule seeds. These seeds are then immobilized on a coverslip via a biotin-neutravidin bridge in a flow chamber.
- Prepare Elongation Mix: Create an elongation buffer containing a low concentration of tubulin, a sustaining concentration of GTP, and a defined fraction of GDP-tubulin (e.g., 0-20%). A key element is including a sufficient concentration of GMPCPP in the mix to suppress catastrophe during the observation window.
- Image Acquisition: Flow the elongation mix into the chamber and immediately begin time-lapse imaging using fluorescence microscopy.
- Data Analysis: Track the growth of both plus and minus ends from the GMPCPP seed over time. Calculate the elongation rates for each end across the different GDP-tubulin conditions.
Expected Outcome: Data supporting the interface-acting model will show a steep, non-linear decrease in the plus-end growth rate as the percentage of GDP-tubulin increases, while the minus-end growth rate will decrease in a more linear, proportional manner.
Protocol 2: Measuring Catastrophe Frequency with Nucleotide Exchange Perturbations
Objective: To test the hypothesis that GDP-to-GTP exchange on terminal microtubule subunits influences catastrophe frequency.
Background: Simulations suggest that allowing GDP-to-GTP exchange on terminal subunits mitigates catastrophe [16]. This can be tested experimentally using reagents that alter nucleotide binding affinity.
Materials:
- Wild-type tubulin
- Mutant tubulin with weakened nucleotide binding affinity [16]
- GTP
- BRB80 buffer
- Flow chamber setup
- TIRF microscope
Method:
- Sample Preparation: Prepare two separate reactions: one with wild-type tubulin and another with mutant tubulin that has a higher rate of nucleotide exchange.
- Dynamic Assay: Immobilize GMPCPP seeds in a flow chamber and initiate growth by flowing in each tubulin preparation in GTP-containing buffer.
- Data Collection: Record movies of microtubule dynamics. For each growing microtubule, measure the time from the start of growth until the moment of catastrophe.
- Quantification: Plot the survival fraction (microtubules that have not yet undergone catastrophe) over time. The catastrophe frequency is inversely related to the time at which 50% of the microtubules have undergone catastrophe.
Expected Outcome: The mutant tubulin with higher nucleotide exchange rates is predicted to exhibit a lower catastrophe frequency compared to wild-type tubulin, supporting a protective role for nucleotide exchange at the microtubule end.
Mechanism and Workflow Visualizations
Troubleshooting Guides
Guide: Microtubule Breakage During Motility Assays
Reported Issue: Microtubules frequently break or completely depolymerize during gliding or motility assays.
Underlying Cause: The mechanical work produced by walking molecular motors can break tubulin dimer interactions, leading to lattice damage and breakage [17]. This is particularly pronounced in GDP-lattice regions, which are less stable than GTP- or taxol-stabilized lattices [17].
Solutions:
- Stabilize the Lattice: Use microtubules stabilized with taxol or assembled with GMPCPP, a non-hydrolysable GTP analogue, to prevent motor-induced destruction [17].
- Modify Buffer Conditions: Include 10% glycerol in your reaction mixture to intrinsically increase microtubule stability [17].
- Reduce Photo-Toxicity: Use Reflection Interference Contrast Microscopy (RICM) to visualize microtubules without the need for fluorescent labels, which can cause photo-damage [17].
- Confirm ATP Dependence: Verify that breakage is ATP-dependent. A lack of breakage in the presence of non-hydrolysable AMPPNP confirms a motor-based mechanism rather than spontaneous lattice disintegration [17].
Guide: Resolving Lattice Heterogeneity in Structural Analysis
Reported Issue: Standard single-particle analysis (SPA) or subtomogram averaging (STA) reveals a seemingly homogenous microtubule lattice, failing to capture structural variations like multiple seams or holes.
Underlying Cause: Conventional averaging techniques combine thousands of images to produce a single representative structure, which masks the intrinsic structural heterogeneity of individual microtubules [18].
Solutions:
- Implement Segmented Averaging: Use a Segmented Subtomogram Averaging (SSTA) strategy. This involves dividing a full-length microtubule model into shorter segments and calculating subtomogram averages for each segment to reveal local variations in seam number and location [18].
- Use Lattice Decorations: Decorate microtubules with kinesin motor-domains, which bind every αβ-tubulin heterodimer. This provides a high-density marker to determine the underlying organization of tubulin dimers [18].
- Model Protofilament Paths: Carefully model individual protofilament paths and microtubule centers in software like IMOD/3dmod to use as a template for sub-volume extraction and analysis in programs like PEET [18].
Frequently Asked Questions (FAQs)
FAQ 1: What is the difference between the A-type and B-type microtubule lattice?
The microtubule lattice is composed of αβ-tubulin heterodymers. The difference lies in their lateral interactions:
- A-type lattice (Heterotypic): Characterized by lateral interactions between α- and β-tubulin monomers (α-β, β-α). This type of interaction is predominantly found at the "seam" of the microtubule [18].
- B-type lattice (Homotypic): Characterized by lateral interactions between like monomers (α-α, β-β). This is the predominant lattice type in microtubules assembled in vitro [18].
FAQ 2: What evidence is there for microtubule self-repair in living cells?
Studies have shown that the microtubule lattice is dynamic and not static. The damage induced by molecular motors walking on the lattice—which can remove tubulin dimers—can be compensated for by the insertion of free tubulin dimers from the cytoplasm back into the lattice shaft. This coupling between motor-based damage and tubulin incorporation is a key self-repair mechanism that allows microtubules to survive mechanical stress [17].
FAQ 3: Why is lattice heterogeneity, such as the presence of multiple seams, significant?
The textbook view of a single seam in a microtubule is an oversimplification. Multi-seams are frequent in microtubules assembled in vitro [18]. Furthermore, the location and number of seams can vary within a single microtubule [18]. These seams can act as pre-existing pathways that accelerate damage propagation and fracture [10]. This heterogeneity has profound consequences for understanding microtubule dynamics, as it suggests tubulin can engage in unique lateral interactions and provides a molecular basis for tubulin exchange not just at the ends, but also in the shaft [18].
FAQ 4: What are the key energetic parameters that govern lattice stability and fracture?
The stability of the GDP-microtubule lattice is governed by the binding energies between tubulin dimers. The total binding energy for a fully surrounded dimer is described as ΔGb = 2ΔGlong + 2ΔGlat [10]. A critical parameter is the lattice anisotropy (A), defined as the ratio of longitudinal to lateral binding energies (A = ΔGlong / ΔG_lat) [10]. Recent research comparing simulations with fracture experiments suggests this intrinsic ratio is bounded at approximately 1.5, indicating a weaker anisotropy than previously predicted from tip-growth models [10].
The following tables consolidate key quantitative findings from recent research on microtubule lattice dynamics.
Table 1: Microtubule Lifetime Under Motor-Induced Stress
| Motor Type | Motor Concentration | Average Microtubule Lifetime (min) | Conditions & Notes |
|---|---|---|---|
| Control (No motor) | - | 20.0 ± 2.0 | RICM, 10% Glycerol [17] |
| Klp2 (Kinesin-14) | 1 nM | 8.7 ± 0.2 | RICM, 10% Glycerol [17] |
| Klp2 (Kinesin-14) | 10 pM | ~20.0 (No significant effect) | RICM, 10% Glycerol [17] |
| Cytoplasmic Dynein | 1 nM | 12.4 (Reduced from 20) | RICM [17] |
| Control (Spontaneous) | - | 12.3 ± 0.1 | Fluorescence microscopy [17] |
| Klp2 (Kinesin-14) | 1 nM | 5.3 ± 0.1 | Fluorescence microscopy [17] |
Table 2: Lattice Fracture Parameters from Kinetic Modeling
| Parameter | Symbol | Value / Range | Significance |
|---|---|---|---|
| Lattice Anisotropy | A = ΔGlong / ΔGlat | ~1.5 (bounded) | Challenges previous predictions; indicates weaker anisotropy [10] |
| Time to Fracture | - | 10 - 20 minutes | For end-stabilized MTs in absence of free tubulin [10] |
| Damage Zone Size | - | ~1 μm | Average length of damaged lattice region before fracture [10] |
| Breaking Event Frequency (Klp2) | - | 1 event / 14 μm | At high motor concentration (10 nM) [17] |
Experimental Protocols
Detailed Protocol: Segmented Subtomogram Averaging (SSTA) for Lattice Analysis
This protocol is used to characterize the structural heterogeneity of individual microtubule lattices, such as the presence and variation of multiple seams [18].
Key Software: IMOD (v4.12.30 or later) and PEET (v1.16.0 alpha or later) [18].
Procedure:
Data and Directory Preparation:
- Obtain a cryo-electron tomogram of your microtubule sample (e.g., from a public repository like EMPIAR).
- Create a working directory and save the tomogram file (e.g.,
your_tomogram.mrc).
Modeling Protofilament Paths with 3dmod:
- Open the tomogram in 3dmod using the command:
3dmod your_tomogram.mrc MT_Model.mod. - In the 3dmod Information Window, select Model mode.
- Use the Image > Slicer function to view the microtubule in cross-section. Adjust the X and Z rotation sliders (e.g., 90.0 and -57.4) to clearly visualize and individualize protofilaments.
- With the Open object type selected and a sphere radius of 3, manually trace the path of a reference protofilament by placing points. It is advised to model consistently from the bottom to the top of the tomogram.
- Regularly save your model during this process.
- Open the tomogram in 3dmod using the command:
Generating the Full Microtubule Model:
- The initial protofilament model serves as a template. Use PEET commands to generate models for all other protofilaments and the microtubule center, creating a full template for the microtubule.
Subtomogram Averaging and Segmentation:
- Extract sub-volumes at every kinesin motor domain position (used for decoration) to compute a full, global subtomogram average of the microtubule.
- Divide this full model into shorter segments (e.g., 100-500 nm in length).
- Using the global average parameters as an initial template, compute a separate subtomogram average for each individual segment.
Analysis of Heterogeneity:
- Analyze the segmented averages to identify local variations, such as changes in the number and position of seams (A-lattice) within the predominantly B-lattice structure.
Detailed Protocol: Assessing Motor-Induced Microtubule Destruction
This protocol describes a "motility assay" to study how walking molecular motors damage and break the microtubule lattice [17].
Key Materials:
- Capped GDP-microtubules (GDP-lattice with GMPCPP-stabilized ends).
- Purified motor proteins (e.g., kinesin-1, kinesin-14/Klp2, or cytoplasmic dynein).
- Flow cells with non-adhesive surface coatings or micropatterned attachments.
Procedure:
Microtubule Preparation:
- Prepare capped GDP-microtubules. This involves growing a GDP-tubulin lattice from a stable seed, but protecting it from spontaneous depolymerization by capping both ends with a short section of GMPCPP-tubulin.
Assay Geometry:
- In a gliding assay, motors are attached to a glass surface, and microtubules glide over them.
- In a motility assay, attach the capped microtubules to the coverslip surface (e.g., via patterned seeds), leaving the majority of the shaft unattached. The motors then walk along these surface-attached microtubules.
Data Acquisition with Reduced Photo-Damage:
- To minimize fluorescence-induced damage, use Reflection Interference Contrast Microscopy (RICM) to visualize label-free microtubules and non-fluorescent motors [17].
- Alternatively, include 10% glycerol in the reaction buffer to increase intrinsic microtubule stability.
Quantification of Breakage:
- Record time-lapse videos of the microtubules after introducing motors and ATP.
- Quantify microtubule lifetime and the frequency of breakage events along the shaft (distinct from the loss of the protective cap).
- Compare these metrics to control experiments performed without motors or without ATP (using AMPPNP).
Signaling Pathways and Workflows
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Microtubule Lattice Dynamics Research
| Reagent / Material | Function / Role | Key Details & Considerations |
|---|---|---|
| GMPCPP Tubulin | Stabilizing GTP analogue | Used to create stable microtubule "caps" or seeds that protect GDP-lattices from end-wise depolymerization; mimics GTP-bound state [17]. |
| Taxol | Microtubule-stabilizing drug | Prevents depolymerization of the entire microtubule in gliding assays, allowing isolation of motor effects on the shaft [17]. |
| Kinesin Motor-Domains | Lattice decoration for structural studies | Binds every αβ-tubulin heterodimer, providing a high-density marker to resolve underlying tubulin organization in techniques like SSTA [18]. |
| Klp2 (Kinesin-14) | Minus-end-directed motor for damage assays | A non-processive motor used in motility assays to study force-induced breakage along the microtubule shaft [17]. |
| Cytoplasmic Dynein | Minus-end-directed motor for damage assays | A processive motor; single dimers can be used at low concentrations (e.g., 50 pM) to induce lattice breakage [17]. |
| AMPPNP | Non-hydrolysable ATP analogue | Locks motors in a bound, non-moving state; used as a negative control to confirm breakage requires motor movement [17]. |
| IMOD/PEET Software | Structural analysis and averaging | Software suite for tomographic reconstruction, modeling, and subtomogram averaging, essential for SSTA [18]. |
| Jak-IN-5 | Jak-IN-5|JAK Inhibitor|C27H31FN6O | |
| TDP1 Inhibitor-1 | TDP1 Inhibitor-1, MF:C26H26N2O5, MW:446.5 g/mol | Chemical Reagent |
Advanced Tools for Lattice Analysis: From Atomic Resolution to Cellular Context
Segmented Subtomogram Averaging (SSTA) for Resolving Lattice Heterogeneity
Purpose and Applications of SSTA
Segmented Subtomogram Averaging (SSTA) is an advanced cryo-electron tomography technique specifically designed to investigate the structural heterogeneity within individual microtubules. Traditional Single-Particle Analysis (SPA) and full-length Subtomogram Averaging (STA) approaches generate averaged structures assumed to represent the entire sample, potentially masking intrinsic variations. In contrast, SSTA addresses this limitation by dividing microtubules into shorter segments for individual analysis, revealing dynamic changes in lattice organization that occur along the shaft of single microtubules [19] [20].
The primary application of SSTA in microtubule research is the detailed characterization of lattice heterogeneity, including:
- Mapping A- and B-lattice seams: Identifying regions where αβ-tubulin heterodimers engage in heterotypic (α-β, β-α) versus homotypic (α-α, β-β) lateral interactions [20]
- Detecting multi-seam configurations: Revealing the presence of multiple seam regions within individual microtubules, contrary to the traditional "textbook" single-seam model [19]
- Identifying lattice discontinuities: Uncovering holes of one to a few tubulin subunits within the microtubule wall that form at transition zones where seam number or location changes [21]
- Comparing assembly conditions: Analyzing structural differences between microtubules assembled from purified tubulin versus those formed in cytoplasmic extracts [21]
This technique has demonstrated that changes in seam number and location are intrinsic properties of microtubules assembled from purified tubulin, and that these transitions occur with varying frequency along individual microtubules [21].
Troubleshooting Guides
Common Experimental Challenges and Solutions
Table 1: Troubleshooting Common SSTA Experimental Issues
| Problem | Possible Causes | Solutions | Prevention Tips |
|---|---|---|---|
| Poor kinesin decoration | Incorrect motor-domain concentration; Non-optimal buffer conditions | Titrate kinesin concentration (typically 1-5 µM); Verify binding conditions using negative stain EM | Always include control microtubules with known decoration pattern; Confirm activity of kinesin stock |
| Low signal-to-noise ratio in tomograms | Insufficient tilts; Radiation damage; Ice contamination | Implement dual-axis tilt series acquisition [21]; Use modern direct electron detectors; Optimize cryo-grid preparation | Use fiduciary gold markers for alignment; Collect data at higher magnification (≥×50,000) for critical regions |
| Incomplete microtubule modeling | Microtubule curvature; High background noise | Model from bottom to top of tomogram; Adjust X-rotation to maximize protofilament contrast [20] | Ensure microtubules are parallel to tomogram plane in thick ice; Use Slicer Window in IMOD for 3D orientation |
| Aberrant protofilaments in averages | Genuine lattice transitions; Registration errors | Apply Segmented SSTA to identify transition zones [21]; Verify with Fourier space filtering | Increase segment length to 125-180 nm for better statistics; Cross-check with raw tomogram data |
Computational and Software Issues
Table 2: Troubleshooting SSTA Computational Workflow
| Issue | Symptoms | Resolution Steps |
|---|---|---|
| PEET/IMOD installation failures | Software won't launch; Missing dependencies | Verify system requirements: macOS, Windows, or Linux with ≥16 GB RAM, 4 physical cores [20]; Check PATH environment variables |
| Model registration failures | Points not aligning to kinesin densities; Poor averages | Adjust sphere radius to 3 in 3dmod Object Type settings [20]; Re-check microtubule center modeling |
| Segment alignment discrepancies | Inconsistent seam counts between segments | Use main parameters of full-length microtubule as template for segments [19]; Verify motive list division |
| Reconstruction artifacts | Streaking; Missing wedges | Implement wedge-masked difference maps [19]; Consider iterative refinement cycles |
Frequently Asked Questions (FAQs)
Q1: What is the key advantage of SSTA over conventional STA for microtubule analysis? A1: While conventional STA generates a single average structure representing the entire microtubule, SSTA divides microtubules into shorter segments (typically 125-180 nm) to analyze structural variations along individual microtubules. This enables detection of changes in seam number and location, as well as identification of lattice discontinuities that would be averaged out in full-length approaches [19] [21].
Q2: Why is kinesin motor-domain decoration essential for SSTA of microtubules? A2: Kinesin motor-domains bind specifically to every β-tubulin subunit within the microtubule lattice, serving as unambiguous markers for determining the underlying organization of αβ-tubulin heterodimers. This binding pattern allows researchers to distinguish between A-lattice (heterotypic) and B-lattice (homotypic) interactions and accurately map seam locations [20] [21].
Q3: How frequently do lattice transitions occur in microtubules? A3: Transition frequency varies significantly between microtubules and assembly conditions. Analysis of microtubules assembled from purified porcine brain tubulin with GTP revealed an average transition frequency of 3.69 µmâ»Â¹, but with high heterogeneity—some microtubules showed no transitions while others reached frequencies up to ~15 µmâ»Â¹ [21].
Q4: Can SSTA be applied to microtubules in intact cells? A4: Currently, application to intact cells is challenging as it requires membrane removal with detergents to allow kinesin access to microtubules. While this has been demonstrated for cytoplasmic microtubules [20], new strategies are needed for studying native microtubule lattices in unperturbed cellular environments.
Q5: What are the minimum computational requirements for SSTA processing? A5: The protocol can be run on computers with at least one CPU with four physical cores and 16 GB of RAM. The software (IMOD and PEET) is multi-platform and compatible with macOS, Windows, and Linux operating systems [20].
Q6: How does buffer condition affect microtubule lattice heterogeneity? A6: The nucleotide conditions (GTP vs. GMPCPP) significantly influence lattice regularity. Microtubules assembled with GMPCPP, a slowly hydrolysable GTP analogue, generally show more regular organization and enable better resolution tomograms due to the ability to use lower tubulin concentrations (10 µM vs. 40 µM for GTP) [21].
Experimental Protocols
Sample Preparation and Data Collection
Microtubule Assembly and Kinesin Decoration:
- Polymerization conditions: Assemble microtubules from purified porcine brain tubulin (10-40 µM) in BRB80 buffer (80 mM PIPES, 1 mM MgCl₂, 1 mM EGTA, pH 6.8) with 1 mM GTP or GMPCPP [21]
- Kinesin decoration: At polymerization plateau, add kinesin motor-domains (final concentration 1-5 µM) and incubate for 5-10 minutes before grid preparation
- Vitrification: Apply 3-5 µL sample to freshly glow-discharged cryo-EM grids, blot, and plunge-freeze in liquid ethane
Cryo-Electron Tomography Data Collection:
- Tilt series acquisition: Collect dual-axis tilt series typically from ±60° with 1-2° increments at 25,000-50,000× magnification [21]
- Defocus range: Use -4 to -8 µm defocus depending on ice thickness and desired resolution
- Dose management: Implement dose-fractionation with total dose of 80-120 eâ»/Ų distributed across the tilt series
SSTA Computational Workflow
Detailed SSTA Processing Steps:
Tomogram Preprocessing (IMOD):
tiltalign: Align tilt series using gold fiducialstomoproc: Apply preprocessing filters and correctionsetomo: Reconstruct tomograms with weighted back-projection or SIRT
Microtubule Modeling (3dmod):
- Open tomogram:
3dmod GMPCPP_tomoFig5_bin4.mrc MT_Model.mod - Use Slicer Window (Image > Slicer) to visualize microtubule cross-sections
- Set Object type to "Open" with Sphere radius for points = 3 [20]
- Model protofilament path by placing points along filament center
- Open tomogram:
Subtomogram Extraction and Averaging (PEET):
- Extract sub-volumes (~50 nm³) at each kinesin motor domain position
- Generate initial averages using full microtubule length
- Divide model into segments of 125-180 nm length
- Process segmented sub-volumes using full-length parameters as template
Heterogeneity Analysis:
- Compare kinesin binding patterns between segments
- Identify lattice transitions by tracking protofilament registration
- Map seam location and count variations along microtubule length
- Detect holes by identifying out-of-phase kinesin periodicity
Table 3: Microtubule Lattice Transition Frequencies Under Different Assembly Conditions
| Assembly Condition | Average Transition Frequency (µmâ»Â¹) | Microtubules Analyzed | Total Segments | Key Observations |
|---|---|---|---|---|
| Purified tubulin + GTP | 3.69 | 24 | 195 | High heterogeneity: 0 to ~15 µmâ»Â¹ transition frequency [21] |
| Purified tubulin + GMPCPP | Reduced (data not quantified) | Multiple | Not specified | More regular organization; clearer hole visualization [21] |
| Xenopus egg cytoplasmic extracts | Lower than in vitro | Multiple | Not specified | Cellular components regulate lattice regularity [21] |
Table 4: Computational Resources and Software Specifications
| Component | Minimum Requirements | Recommended Specifications |
|---|---|---|
| Computer Hardware | 4 physical cores, 16 GB RAM | Apple M1 Max 3.22 GHz, 64 GB RAM, 4 TB SSD [20] |
| Software Versions | IMOD 4.12.30, PEET 1.16.0 alpha | Latest stable releases with bug fixes |
| Peripheral Equipment | Standard three-button mouse | Extended keyboard with numeric keypad [20] |
| Storage | 500 GB free space | 4 TB SSD for large datasets |
Research Reagent Solutions
Table 5: Essential Research Reagents for SSTA Experiments
| Reagent / Material | Specifications | Function in SSTA Workflow |
|---|---|---|
| Porcine brain tubulin | Purified (>95% purity), 10-40 µM working concentration | Microtubule polymerization substrate for in vitro assembly [21] |
| Kinesin motor-domains | Recombinantly expressed, purified, 1-5 µM decorating concentration | Binds every αβ-tubulin heterodimer to mark lattice organization [19] [20] |
| GMPCPP | Slowly hydrolysable GTP analogue, 1 mM in assembly buffer | Produces more stable, regular microtubules for high-resolution analysis [21] |
| Cryo-EM grids | Quantifoil or C-flat, freshly glow-discharged | Sample support for vitrification and tomography data collection |
| Xenopus egg cytoplasmic extracts | Freshly prepared, competence-verified | Physiological microtubule assembly environment comparison [21] |
Kinetic Monte Carlo Simulations for Modeling Lattice Dynamics and Fracture
Frequently Asked Questions: Kinetic Monte Carlo Method
Q1: What is the fundamental principle behind Kinetic Monte Carlo (KMC) simulations? KMC is a stochastic simulation technique designed to model the time evolution of systems dominated by rare events. Unlike molecular dynamics, which simulates every vibration, KMC jumps from state to state, with the probability of each transition governed by its rate constant. This makes it particularly suited for simulating processes like microtubule lattice dynamics and fracture, which occur on timescales far beyond those accessible by atomistic simulation [22] [23].
Q2: In the context of microtubule fracture, what constitutes an "event" in a KMC simulation? An "event" is the discrete detachment of a tubulin dimer from the lattice. The rate of detachment ((k{mn})) for a specific dimer depends on the number of its intact longitudinal ((m)) and lateral ((n)) monomer contacts, following the Arrhenius-type equation [10]: [ k{mn} = \frac{1}{\tau}e^{\beta\left(m\Delta G{\text{long}} + {n\over 2}\Delta G{\text{lat}} - {\Delta G{\text{b}}\over 2}\right)} ] Here, (1/\tau) is the off-rate constant for a corner dimer, (\beta) is the inverse thermodynamic temperature, and (\Delta G{\text{long}}) and (\Delta G_{\text{lat}}) are the longitudinal and lateral binding energies, respectively [10].
Q3: Our KMC simulation of microtubule fracture is running too slowly. What could be the cause?
KMC simulations can be slowed down by a "timescale disparity problem," where one type of event (e.g., dimer detachment) is extremely rare compared to others. Furthermore, in a lattice model, the algorithm must check the status of every dimer and its neighbors at every step. For a large 10µm microtubule containing hundreds of thousands of dimers, this computational overhead is significant. Implementing advanced KMC algorithms, like the (n)-fold way or using performance-optimized libraries such as kmos, can help overcome this [22] [23].
Q4: Why does a monomer vacancy in the microtubule lattice lead to asymmetric fracture propagation? A monomer vacancy creates a local defect. When the simulation starts from a monomer vacancy within the B-lattice, two new seams emanate from the defect. As the vacancy grows, the longitudinal fracture front that propagates across these seams gains "seam dimers" with different neighbor counts, accelerating its detachment rate compared to the front propagating into the perfect B-lattice. This breaks the symmetry and leads to faster propagation in one direction [10].
Q5: How is lattice anisotropy defined and what is its significance in microtubule fracture models? Lattice anisotropy ((A)) is defined as the ratio of longitudinal to lateral binding energies [10]: [ A = \frac{\Delta G{\text{long}}}{\Delta G{\text{lat}}} ] This parameter critically influences the shape and speed of damage propagation. Recent KMC studies matching experimental fracture data suggest the intrinsic anisotropy is bounded at approximately 1.5, which is lower than previous estimates from tip-growth models. This weaker anisotropy results in more balanced longitudinal and lateral crack growth [10].
Troubleshooting Guides
Issue 1: Simulation Does Not Reproduce Experimental Fracture Times
Problem The simulated time for a microtubule to fracture is significantly shorter or longer than the experimentally observed 10-20 minutes [10].
Investigation & Resolution
| Investigation Step | Action & Parameters to Check |
|---|---|
| Verify Lattice Energy Parameters | Confirm the values for (\Delta G{\text{long}}) and (\Delta G{\text{lat}}) in your input file. The total binding energy for a fully surrounded dimer is (\Delta Gb = 2\Delta G{\text{long}} + 2\Delta G_{\text{lat}}), and it must be strongly negative ((\ll kT)) to ensure a stable lattice [10]. |
| Check the Anisotropy Ratio | Ensure the anisotropy (A = \Delta G{\text{long}} / \Delta G{\text{lat}}) is set appropriately. For GDP microtubules, a value around 1.5 is consistent with recent fracture data [10]. |
| Inspect Initial Defect Setup | Validate the initial condition. The simulation should start from a defined defect (e.g., a monomer vacancy) rather than a perfect lattice, as spontaneous dimer loss from a full lattice is a rare event [10]. |
Issue 2: Unphysical Fracture Propagation Patterns
Problem The damage in the microtubule lattice grows in a geometrically irregular or unexpected shape that does not align with theoretical predictions.
Investigation & Resolution
| Investigation Step | Action & Parameters to Check |
|---|---|
| Validate Detachment Rates | Review the calculated rates for all possible neighbor configurations ((k{mn})). A dimer's detachment rate increases as it loses neighbors. The rate for a corner dimer ((k{12})) should be the fastest [10]. |
| Account for Seam Dynamics | Check if your model correctly handles seams (lattice defects where α-tubulin meets β-tubulin). A dimer on a seam has three lateral neighbors instead of two, which increases its detachment rate ((k{13} > k{14})) and accelerates fracture propagation along that boundary [10]. |
| Confirm Boundary Conditions | For simulating bulk lattice fracture, ensure both ends of the microtubule are stabilized with a non-detachable cap, preventing depolymerization from the ends from confounding the results [10]. |
Issue 3: Handling Lateral Interactions in the Lattice Model
Problem The model fails to capture collective dynamics or produces results that deviate from mean-field approximations.
Investigation & Resolution
- Process Inclusion: Ensure all relevant elementary processes are included. A model that only includes dimer detachment may be insufficient if reattachment or other local rearrangements are significant in your system [22].
- Lateral Interactions: Confirm whether your model needs to account for explicit lateral interactions between dimers beyond simple neighbor counting. Omitting these can lead to errors in simulating collective behavior [22].
- Code Verification: Use a well-established KMC framework like
kmosto benchmark your custom implementation against a standardized code [22].
Experimental Protocols & Data
Protocol: KMC Simulation of Microtubule Fracture from a Monomer Vacancy
This protocol details how to set up a KMC simulation to model fracture propagation from an initial monomer vacancy in a stabilized GDP-microtubule, based on the methodology from recent research [10].
1. System Setup and Lattice Definition
- Construct a two-dimensional lattice representing 13 protofilaments.
- Define the lattice length (e.g., corresponding to a 10 µm microtubule).
- Assign α- and β-tubulin monomers to their positions, identifying the single seam of the B-lattice and any additional seams arising from defects.
- Stabilize both ends of the microtubule by defining the first and last few dimers as non-detachable.
2. Parameter Initialization Populate the following table with the required energy and rate parameters:
| Parameter | Symbol | Value (Example) | Description |
|---|---|---|---|
| Longitudinal Binding Energy | (\Delta G_{\text{long}}) | - | Energy per longitudinal monomer contact [10]. |
| Lateral Binding Energy | (\Delta G_{\text{lat}}) | - | Energy per lateral monomer-monomer contact [10]. |
| Lattice Anisotropy | (A) | ~1.5 | Ratio (\Delta G{\text{long}} / \Delta G{\text{lat}}) [10]. |
| Inverse Temperature | (\beta) | (1/kT) | Inverse thermodynamic temperature [10]. |
| Fundamental Time Constant | (\tau) | - | Time constant related to the off-rate of a corner dimer [10]. |
| Monomer Size | (\eta) | 4 nm | Size of a single tubulin monomer [10]. |
3. Initialization and Execution
- Introduce a single monomer vacancy at the desired location (e.g., within the B-lattice, or adjacent to a seam).
- At each KMC step:
- Catalog Events: Identify all dimers on the boundary of any vacancy and calculate their individual detachment rates (k{mn}) using Equation (3).
- Select Event: Calculate the total rate (R{\text{tot}} = \sum k{mn}). Choose a event with probability (k{mn}/R{\text{tot}}) using a random number.
- Advance Time: Increment the simulation clock by (\Delta t = -\ln(r)/R{\text{tot}}), where (r) is a uniform random number between 0 and 1.
- Update Lattice: Execute the chosen dimer detachment event, updating the lattice structure and the list of available events.
- Continue the loop until the damage spans all 13 protofilaments, marking complete fracture.
Quantitative Data from Microtubule Fracture Studies
The following table summarizes key quantitative findings from KMC simulations and related experiments on microtubule lattice fracture [10].
| Observable / Parameter | Experimental Finding | KMC Simulation Insight |
|---|---|---|
| Time to Fracture | 10 to 20 minutes in end-stabilized MTs [10]. | Reproduced with specific (\Delta G{\text{long}}) and (\Delta G{\text{lat}}) values. |
| Damage Size at Fracture | Averages about 1 µm along the MT axis [10]. | Determined by the relative speeds of longitudinal vs. lateral crack growth. |
| Lattice Anisotropy ((A)) | Not directly measurable. | Best fit to fracture data suggests (A \approx 1.5), weaker than prior estimates [10]. |
| Effect of Monomer Vacancy | Linked to lattice instability [10]. | Acts as nucleation site for fracture; leads to asymmetric propagation in multi-seam structures [10]. |
| Seam Dimer Detachment Rate | Not directly measurable. | (k{13} > k{14}), leading to accelerated longitudinal propagation along seams [10]. |
Model Visualization
KMC Algorithm Workflow
Microtubule Fracture Propagation from a Defect
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in Context of KMC Microtubule Studies |
|---|---|
| End-Stabilizing Caps (e.g., GMPCPP, Taxol) | Used experimentally to create a stable microtubule seed and to prevent depolymerization from the ends. This allows for the isolated study of lattice fracture dynamics away from the tips [10]. |
| Lattice Energy Parameters ((\Delta G{\text{long}}, \Delta G{\text{lat}})) | These are not physical reagents but critical input parameters for the KMC model. They are derived by fitting simulation results (e.g., fracture time) to experimental data [10]. |
KMC Simulation Framework (e.g., kmos) |
A software platform that facilitates the implementation of lattice-based KMC models, helping to manage the catalog of events and ensure correct algorithm execution [22]. |
| Sniper(abl)-020 | Sniper(abl)-020, MF:C44H59ClN10O8S, MW:923.5 g/mol |
| Zotatifin | Zotatifin, CAS:2098191-53-6, MF:C28H29N3O5, MW:487.5 g/mol |
Super-Resolution Microscopy for Nanoscale Visualization of Drug-Induced Dysfunction
This technical support guide is designed for researchers characterizing drug-induced microtubule lattice dynamics. Super-resolution microscopy enables the visualization of nanoscale microtubule dysfunction, such as the increased filament curvature and fragmentation caused by antimitotic drugs like colcemid at concentrations as low as 30-80 nM [24]. However, achieving consistent, high-quality results requires careful optimization of imaging techniques, sample preparation, and data analysis. The following sections provide targeted troubleshooting guidance and detailed protocols to address the most common technical challenges in this specialized research area.
Frequently Asked Questions (FAQs)
Q1: My super-resolution images of drug-treated microtubules appear blurry and lack the expected resolution improvement. What are the primary factors I should check?
A: Blurry super-resolution images typically stem from three main areas: sample preparation, fluorophore selection, or imaging parameter optimization. First, verify that your fluorescent probes are specifically targeting microtubules with high signal-to-noise ratio. For STORM imaging, ensure your blinking buffer is fresh and properly optimized to achieve sparse, stochastic activation of fluorophores. For live-cell imaging, confirm that your cellular health is maintained throughout the experiment, as drug treatments can compromise cell viability and introduce artifacts [25].
Q2: What is the optimal super-resolution technique for visualizing nanoscale drug-induced damage in microtubule lattice structure?
A: The choice depends on your specific research question and whether you need live-cell or fixed-cell imaging. For fixed-cell studies of nanoscale microtubule curvature and fragmentation, dSTORM provides exceptional resolution (20-30 nm) and is well-suited for quantifying structural abnormalities [24]. For live-cell imaging of microtubule dynamics in response to drug treatments, STED or SOFI offer better temporal resolution while still providing super-resolution capability [24] [26]. If you need to track drug distribution simultaneously with structural changes, consider using SIM with specifically designed molecular probes [25].
Q3: How can I quantitatively measure drug-induced microtubule curvature and fragmentation from my super-resolution data?
A: Quantifying these parameters requires specialized analysis approaches. For curvature measurement, implement an algorithm that fits splines to the microtubule filaments and calculates the radius of curvature along their length. Research indicates that curvatures greater than 2 rad/μm are associated with microtubule breakage [24]. For fragmentation analysis, develop a pipeline that identifies individual microtubule filaments, detects discontinuities, and quantifies fragment length distribution. Ensure your analysis accounts for the possibility of different damage phenotypes depending on drug concentration and exposure time [24].
Q4: What controls are essential for validating that observed microtubule dysfunction is specifically drug-induced?
A: Implement a comprehensive control strategy including: (1) Untreated cells to establish baseline microtubule architecture; (2) Vehicle-only treated cells to control for solvent effects; (3) Concentration gradient of the drug to demonstrate dose-dependence; (4) Time-course experiments to track the progression of damage; (5) If available, use of microtubule-stabilizing agents in combination with your drug to confirm specificity of effect [24] [3].
Troubleshooting Guides
Common Technical Issues and Solutions
Table 1: Troubleshooting Guide for Super-Resolution Imaging of Drug-Treated Microtubules
| Problem | Potential Causes | Solutions | Preventive Measures |
|---|---|---|---|
| Poor signal-to-noise ratio | Fluorophore bleaching; Inadequate labeling; Drug-induced autofluorescence | Optimize dye concentration; Use antifade reagents; Test different filter sets | Validate probes in untreated cells first; Use fresh imaging buffers [25] |
| Non-specific probe labeling | Probe concentration too high; Inadequate washing; Drug affecting cellular uptake | Titrate probe concentration; Increase wash steps; Include control without primary antibody | Characterize drug-probe interactions in vitro before cellular experiments [25] |
| Excessive background in STORM/dSTORM | Inadequate blinking buffer; Oxygen scavenging system failure; Sample too thick | Freshly prepare blinking buffer; Check glucose oxidase/catalase activity; Use thinner samples | Aliquot blinking buffer components; Validate system with test samples [26] |
| Cell morphology changes during live imaging | Drug toxicity; Phototoxicity; Environmental control failure | Optimize drug concentration; Reduce laser power; Verify incubation system stability | Monitor cell health indicators pre-imaging; Use lower drug doses for longer times [24] |
| Inconsistent drug effects across replicates | Variable drug solubility; Cell confluence differences; Microtubule stabilization state | Include drug solubility controls; Standardize cell culture conditions; Control for passage number | Pre-make drug aliquots at consistent concentrations; Document cell culture parameters [24] [3] |
Quantitative Analysis of Drug Effects
Table 2: Quantifiable Parameters for Drug-Induced Microtubule Dysfunction
| Parameter | Measurement Method | Typical Control Values | Drug-Induced Changes | Technical Considerations |
|---|---|---|---|---|
| Curvature (rad/μm) | Spline fitting along filament axis | <0.5 rad/μm [24] | ≥2.0 rad/μm indicates damage progression [24] | Ensure sufficient point density for accurate fitting |
| Fragmentation index | Number of breaks per μm microtubule | <0.05 breaks/μm | ≥0.15 breaks/μm with ≥100 nM colcemid [24] | Distinguish true breaks from out-of-focus regions |
| Lattice spacing changes | Cryo-EM correlation; MAP binding patterns | 81.7±0.1Å (compacted) to 83.5±0.2Å (expanded) [3] | Drug-dependent expansion or compaction [3] | Requires complementary techniques for verification |
| Damage propagation rate | Time-lapse measurement of defect expansion | Minimal in untreated cells | 10-20 minutes to fracture from initial defect [10] | Critical for live-cell imaging experiments |
| Seam involvement in damage | Lattice orientation mapping | Seams as inherent structural features | Seams accelerate damage propagation [10] | Requires high-resolution techniques with seam discrimination |
Experimental Protocols
Sample Preparation for Microtubule Super-Resolution Imaging
Protocol 1: Fixed-Cell Preparation for Microtubule dSTORM Imaging
This protocol optimizes preservation of drug-induced microtubule structures for super-resolution analysis.
Cell Culture and Drug Treatment:
- Plate cells on high-precision #1.5H glass-bottom dishes 24 hours before experiment
- Treat with antimitotic drug (e.g., colcemid at 30-100 nM) for predetermined time [24]
- Include vehicle-only controls for each experiment
Fixation:
- Aspirate medium and rinse briefly with pre-warmed PBS-MT (PBS + 1 mM MgClâ‚‚ + 0.5 mM CaClâ‚‚)
- Fix with 3% formaldehyde + 0.1% glutaraldehyde in PBS-MT for 15 minutes at 37°C
- Quench with 0.1% sodium borohydride in PBS for 7 minutes
Immunostaining:
- Permeabilize with 0.5% Triton X-100 in PBS for 10 minutes
- Block with 5% BSA + 0.1% fish skin gelatin in PBS for 1 hour
- Incubate with primary anti-tubulin antibody (1:200) overnight at 4°C
- Wash 3× with PBS over 30 minutes
- Incubate with photoswitchable secondary antibody (e.g., Alexa Fluor 647) for 1 hour at room temperature
- Wash 3× with PBS over 30 minutes
Imaging Buffer Preparation (for dSTORM):
Super-Resolution Imaging Workflow
Protocol 2: dSTORM Imaging of Drug-Induced Microtubule Damage
This protocol details the acquisition parameters for visualizing nanoscale microtubule dysfunction.
System Setup:
- Use TIRF or highly inclined illumination for optimal signal-to-noise
- Ensure stable temperature control at 20°C during imaging
- Verify laser alignment and calibration with fluorescent beads
Image Acquisition:
- Acquire widefield image to locate cells and assess overall microtubule structure
- Switch to dSTORM imaging mode with 640 nm activation laser
- Acquire 10,000-20,000 frames at 50-100 ms exposure time
- Ensure appropriate fluorophore blinking density (0.5-1 molecules/μm²)
Data Reconstruction:
Diagram 1: Experimental workflow for super-resolution imaging of drug-induced microtubule dysfunction, showing the three main phases of sample preparation, imaging, and data analysis.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Microtubule Super-Resolution Studies
| Category | Specific Item | Function/Application | Technical Notes |
|---|---|---|---|
| Microtubule Drugs | Colcemid | Induces microtubule curvature and fragmentation at nanomolar concentrations (30-100 nM) [24] | Use fresh stock solutions; Concentration-dependent effects |
| Paclitaxel (Taxol) | Microtubule stabilizer; Expands lattice spacing [3] | Useful as control and for lattice spacing studies | |
| Super-Resolution Probes | Alexa Fluor 647 | Photoswitchable dye for dSTORM; High photon yield | Ideal for fixed-cell imaging; Pair with appropriate secondary antibody |
| DNA-PAINT compatible probes | Creates transient binding for localization | Reduced photobleaching; More complex sample preparation | |
| Microtubule Labels | Anti-α-tubulin antibodies | Primary recognition for immunostaining | Validate species specificity and lot-to-lot consistency |
| Live-cell tubulin probes (SIR-tubulin) | For dynamic studies of microtubule dysfunction | Higher background but enables live-cell imaging | |
| Imaging Buffers | dSTORM blinking buffer | Enables stochastic photoswitching | Critical for single-molecule localization; Prepare fresh |
| Live-cell imaging media | Maintains cell health during time-lapse | pH stabilization; Low background fluorescence | |
| Specialized Equipment | TIRF microscope | Creates evanescent field for reduced background | Essential for high-quality single-molecule imaging |
| High-numerical aperture objectives | Maximizes photon collection | ≥1.49 NA recommended for best resolution | |
| Analysis Software | Localization software (ThunderSTORM) | Reconstructs super-resolution images from single-molecule data | Open-source options available; Customize for microtubule analysis |
| Curvature analysis algorithms | Quantifies microtubule bending from drug effects | Implement using MATLAB, Python, or ImageJ plugins | |
| SOCE inhibitor 1 | SOCE inhibitor 1, MF:C25H22F3N5O4, MW:513.5 g/mol | Chemical Reagent | Bench Chemicals |
| URAT1 inhibitor 1 | URAT1 inhibitor 1, MF:C19H15Br2N5O2S2, MW:569.3 g/mol | Chemical Reagent | Bench Chemicals |
Diagram 2: Mechanism of drug-induced microtubule dysfunction showing parallel pathways of structural curvature and lattice fracture, highlighting the role of seams in accelerating damage.
FAQs and Troubleshooting Guides
Troubleshooting Common Experimental Challenges
| Problem Area | Specific Issue | Potential Causes | Recommended Solutions |
|---|---|---|---|
| Assay Signal & Background | High background signal | Inadequate plate washing; insufficient blocking; non-specific antibody binding; contaminated buffers [27]. | Increase number of wash steps; optimize blocking buffer concentration/time; use fresh reagents; check antibody specificity [27]. |
| Weak or no signal | Incorrect incubation times/temperatures; degraded or incorrect reagents; analyte concentration below detection limit; pipetting errors [27]. | Verify protocol incubation steps; use fresh, properly stored standards/antibodies; concentrate or less dilute sample; check pipette calibration [27]. | |
| Data Quality & Reproducibility | High coefficient of variance (CV) between replicates | Pipetting errors; inconsistent reagent mixing; bubbles in wells; inadequate or inconsistent plate washing [27]. | Calibrate pipettes; mix all samples/reagents thoroughly; pop bubbles before reading; ensure consistent washing across all wells [27]. |
| Inconsistent results across plate | Plates stacked during incubation; wells drying out; dirty plate bottom [28]. | Avoid stacking plates during incubations; do not leave plates unattended after washing; clean bottom of plate before reading [28]. | |
| Surface Preparation & Functionalization | Non-specific binding in TIRF/IRM assays | Improperly cleaned or silanized glass surfaces [29]. | Follow rigorous cleaning protocols (e.g., sonication in KOH, ethanol); validate silanization by checking water contact angle >90° [29]. |
Frequently Asked Questions
Q1: My standard curve is poor or doesn't fit well. What should I check?
- Cause: This is often due to an incorrect standard solution, a degraded standard, or using the wrong curve-fitting model [27].
- Solution: Double-check the concentration and dilution calculations of your standard stock. Ensure lyophilized standards are reconstituted exactly as instructed. Verify that the standard has not expired and has been stored correctly. Use the curve-fitting model (e.g., 4- or 5-parameter logistic) recommended by the kit manufacturer [27].
Q2: How can I prevent non-specific binding of proteins in my microtubule imaging assay?
- Cause: Insufficient blocking or non-optimized surface chemistry can lead to proteins sticking to surfaces other than the intended binding sites [29] [27].
- Solution: For glass surfaces used in microscopy (TIRF/IRM), a thorough cleaning and silanization protocol is crucial to create a uniform hydrophobic surface [29]. In immunoassays, ensure the blocking step is optimized; consider increasing the concentration of the blocking agent, extending the incubation time, or adding a small amount of non-ionic detergent like Tween-20 to the buffer [27].
Q3: Why am I seeing inconsistent absorbance readings across my ELISA plate?
- Cause: This is frequently a result of uneven temperature during incubation, inconsistent pipetting, or inadequate washing [28].
- Solution: Avoid stacking plates during incubations to ensure even temperature distribution. Ensure pipettes are properly calibrated and that tips are securely attached. Take care to pipette reagents against the side of the well and avoid bubbles. Make sure the plate washer is functioning correctly and that all wells are washed equally [28] [27].
Experimental Protocols and Workflows
Protocol 1: Microtubule Dynamics Assay Using TIRF Microscopy
This protocol is used to visualize and characterize the dynamic instability of microtubules in a minimal system, crucial for studying intrinsic tubulin properties and the effects of MAPs or drugs [29] [30].
1. Preparation of Glass Surfaces:
- Cleaning: Place glass coverslips in a rack and sonicate in 3M KOH for 20 minutes. Rinse 5x with ultra-pure water, then sonicate again in water and 100% ethanol for 20 minutes each. Dry with compressed air or nitrogen [29].
- Silanization: Submerge the cleaned coverslips in a solution of 0.54M imidazole in acetonitrile. Add trimethylchlorosilane (TMCS) to a final concentration of 3.3% (v/v) and incubate at 45°C for 3 hours. Wash by sonicating in fresh methanol and then rinse with water. Dry in a dust-free environment [29].
- Validation: Validate silanization by applying a 10 µL water droplet to the coverslip. A sufficient hydrophobicity is indicated by a water contact angle greater than 90° [29].
2. Assembling the Flow Chamber and Immobilizing Microtubule Seeds:
- Construct a flow chamber using the silanized coverslip and a glass slide separated by double-sided tape.
- Flow in neutravidin or an anti-rhodamine antibody (depending on your seed labeling) to coat the hydrophobic surface.
- Flow in biotinylated or rhodamine-labeled, GMPCPP-stabilized microtubule "seeds" and allow them to immobilize on the surface [29].
3. Imaging Microtubule Dynamics:
- Prepare a tubulin reaction mix containing purified tubulin (a fraction of which is fluorescently labeled), an oxygen scavenger system (to reduce photobleaching), and a nucleotide (typically GTP) in a suitable buffer.
- Flow the reaction mix into the chamber.
- Image the growth of microtubules from the immobilized seeds using a TIRF microscope. The fluorescent tubulin incorporates into the growing microtubule, allowing visualization of dynamics in real-time [29].
Protocol 2: Reconstitution of ECF Transporters in Proteoliposomes
This protocol describes how to reconstitute integral membrane transporters into liposomes to study their kinetic behavior and competition in a minimal, controlled lipid environment [31].
1. Protein Purification:
- Overproduce and purify the complete ECF transporter complexes (e.g., ECF-FolT2 for folate and ECF-PanT for pantothenate) from your chosen expression system. The ECF module and S-components can be purified together as a complex or as solitary subunits [31].
2. Reconstitution into Proteoliposomes:
- Prepare liposomes from purified lipids using standard methods like extrusion.
- Mix the purified ECF transporters with the pre-formed liposomes in the presence of a detergent.
- Remove the detergent (e.g., via dialysis or bio-beads) to allow the proteins to incorporate into the lipid bilayer, forming sealed proteoliposomes [31].
3. Active Transport Assay:
- Prepare proteoliposomes with the desired substrate (e.g., radiolabeled folate or pantothenate) present outside the liposomes and Mg²âº-ATP present inside the liposomes.
- Initiate transport by adding the proteoliposomes to the external substrate solution.
- At timed intervals, remove aliquots and separate the proteoliposomes from the external medium (e.g., by filtration).
- Quantify the amount of substrate taken up into the proteoliposome lumen, for example, by measuring radioactivity. Active, ATP-dependent transport will show accumulation over time [31].
The Scientist's Toolkit
Key Research Reagent Solutions
| Item | Function / Application | Key Details / Considerations |
|---|---|---|
| Tubulin | Core protein for polymerizing microtubules [29] [3]. | Can be purified from brain tissue or recombinant sources; quality affects assembly competence; can be fluorescently labeled for TIRFM [29]. |
| Paclitaxel (Taxol) | Microtubule-stabilizing drug; a "microtubule expander" [3]. | Nucleates expanded microtubule lattices; used to study lattice spacing and as a control for stabilized structures [3]. |
| Doublecortin (DCX) | Neuronal Microtubule-Associated Protein (MAP); a "microtubule compactor" [3]. | Binds preferentially to and stabilizes compacted microtubule lattices; used in competition studies with expanders like paclitaxel [3]. |
| GMPCPP | Non-hydrolyzable GTP analog [3]. | Used to form stable, non-dynamic microtubule "seeds" from which dynamic GDP-microtubules can grow in assays [3]. |
| Trimethylchlorosilane (TMCS) | Silanizing agent for glass surfaces [29]. | Renders glass hydrophobic for stable adsorption of linker proteins (e.g., neutravidin) in flow chamber assays [29]. |
| Neutravidin | Linker protein for surface functionalization [29]. | Adsorbs to silanized glass; used to immobilize biotinylated microtubule seeds in TIRF assays [29]. |
| S-components (e.g., FolT2, PanT) | Substrate-binding subunits of ECF transporters [31]. | Determine substrate specificity; can dynamically associate/dissociate from the core ECF module in group II transporters [31]. |
| ECF Module (EcfA, EcfA', EcfT) | Energy-coupling core of ECF transporters [31]. | Heterodimeric ATPases (EcfA/A') with a transmembrane subunit (EcfT); provides energy for transport via ATP hydrolysis [31]. |
| bio-THZ1 | bio-THZ1, MF:C52H65ClN12O8S, MW:1053.7 g/mol | Chemical Reagent |
| Shmt-IN-1 | Shmt-IN-1, MF:C18H16Cl2N4O, MW:375.2 g/mol | Chemical Reagent |
Kinetic Parameters of ECF Transporters in Proteoliposomes
The table below summarizes key transport parameters for reconstituted ECF transporters, which can be derived from initial rate of uptake measurements [31].
| Transporter | Substrate | Apparent Km (Substrate) | Km (ATP) | Key Finding |
|---|---|---|---|---|
| ECF-PanT | Pantothenate | Nanomolar range [31] | mM range [31] | Hyperbolic kinetics, indicating a single binding site [31]. |
| ECF-FolT2 | Folate | Nanomolar range [31] | mM range [31] | Hyperbolic kinetics, indicating a single binding site [31]. |
Competition Dynamics in Microtubule Lattice Spacing
This table conceptualizes the competitive interaction between a microtubule expander and compactor, a phenomenon observable in vitro and in cells [3].
| Relative Concentration (DCX : Paclitaxel) | Observed Effect on Microtubule Lattice | DCX Localization Pattern in Cells |
|---|---|---|
| Low DCX, High Paclitaxel | Lattice is expanded [3]. | DCX relocalizes to curved, ring-like segments [3]. |
| High DCX, High Paclitaxel | Lattice is compacted [3]. | DCX binds along straight microtubule sections [3]. |
| Balanced Concentrations | Mixed/complex lattice states [3]. | DCX preferentially binds to a subset of long, straight sections alongside short bends [3]. |
FAQs and Troubleshooting Guides
Foundational Concepts
Q1: What is the difference between microtubule "tip dynamics" and "lattice dynamics"? Microtubules exhibit two key dynamic behaviors. Tip dynamics (or dynamic instability) refers to the stochastic growth and shrinkage occurring at microtubule ends, which is essential for functions like chromosome segregation [32]. Lattice dynamics refers to the continuous, energy-independent exchange and rearrangement of tubulin dimers within the microtubule shaft itself, which facilitates self-repair and resilience against mechanical stress [4]. These processes are interconnected through the conformational states of tubulin subunits [32].
Q2: What are "expanded" and "compacted" microtubule lattice states? The microtubule lattice is structurally plastic and can exist in different conformational states, primarily distinguished by their lattice spacing (dimer rise):
- Expanded State: Has a lattice spacing of approximately 83.5 Ã…. This state is often associated with GTP-tubulin or microtubules bound to proteins like kinesin-1 or drugs like paclitaxel [3].
- Compacted State: Has a lattice spacing of approximately 81.7 Ã… (a 2.3% difference). This state is often linked to GDP-tubulin and is favored by MAPs like doublecortin (DCX) [3]. The transition between these states is central to microtubule mechanics, regulation by MAPs, and the "tubulin code" [3].
Assay Troubleshooting
Q3: My in vivo microtubule/tubulin ratio assay shows high background or poor separation. What could be wrong? This common issue in fractionation-based kits (like BK038 from Cytoskeleton Inc.) often stems from the lysis and stabilization steps [33].
- Cause 1: Ineffective Microtubule Stabilization. Microtubules are depolymerized during lysis. Ensure the lysis/stabilization buffer contains protease inhibitors and is used ice-cold to preserve the native polymerized state.
- Cause 2: Incomplete Centrifugation. The ultracentrifugation step (100,000× g) is critical for cleanly separating polymerized microtubules (pellet) from free tubulin (supernatant). Verify your centrifuge's calibration and use the correct rotor and tubes.
- Cause 3: Microtubule Disassembly During Processing. Keep samples at 37°C after lysis and during centrifugation to prevent cold-induced depolymerization [33].
Q4: In my in vitro binding assay, the protein of interest does not co-sediment with microtubules. What should I check? This indicates a lack of binding in the co-sedimentation assay [34].
- Solution 1: Check Protein Stability. Centrifuge your protein without microtubules first. If it pellets alone, it may be aggregating, which will confound results.
- Solution 2: Optimize Buffer Conditions. High salt concentrations (e.g., >60 mM NaCl) can disrupt electrostatic interactions necessary for many MAP-microtubule bindings. Use low-salt buffers like BRB80.
- Solution 3: Validate Microtubule Integrity and Concentration. Use fresh, taxol-stabilized microtubules polymerized from high-purity tubulin. Ensure the microtubule concentration is sufficient to detect binding [34].
- Solution 4: Include Rigorous Controls. Always run parallel assays with a known non-binding protein (e.g., BSA) and a known MAP-positive control (e.g., a MAP fraction) to validate your experimental setup [34].
Q5: How does tau protein, a known microtubule stabilizer, paradoxically accelerate tubulin exchange in the lattice? Recent research reveals that tau is not just a passive stabilizer. While it stabilizes longitudinal tubulin-tubulin contacts (along the protofilament), it appears to destabilize lateral contacts (between protofilaments) [4]. This increase in lattice anisotropy promotes the mobility of topological defects, facilitating the removal of damaged tubulin dimers and their replacement with new ones. This exchange occurs predominantly at lattice defect sites and represents an active, tau-mediated self-repair mechanism [4].
Table 1: Key Metrics in Tau-Mediated Lattice Dynamics
| Parameter | Condition (15 min incubation) | Value | Biological Significance |
|---|---|---|---|
| Tubulin Incorporation Length | No Tau | 0.7 µm | Baseline, intrinsic lattice dynamics [4] |
| 20 nM Tau | 1.2 µm | Tau enhances longitudinal incorporation [4] | |
| Spatial Frequency of Incorporation | No Tau | 12.4 µm | Lower density of incorporation sites [4] |
| 20 nM Tau | 6.6 µm | Tau increases the frequency of incorporation events [4] | |
| Overall Tubulin Incorporation | No Tau | 1x (Baseline) | -- |
| 20 nM Tau | ~4x Increase | Major enhancement of lattice turnover [4] | |
| Lattice Fluorescence Reduction (at incorporation sites) | 15 min incubation | ~7-15% | Indicates tubulin exchange, not just addition [4] |
Table 2: Microtubule Lattice Spacing States and Regulators
| Lattice State | Lattice Spacing (Dimer Rise) | Associated Nucleotide | Example Preferring Factors |
|---|---|---|---|
| Expanded | 83.5 ± 0.2 Å | GTP-like | Paclitaxel, Kinesin-1, TPX2 [3] |
| Compacted | 81.7 ± 0.1 Å | GDP | Doublecortin (DCX), EB proteins, Tau envelopes [3] |
Detailed Experimental Protocols
Protocol 1: In Vivo Microtubule/Tubulin Ratio Assay
This protocol separates and quantifies polymerized microtubules from free tubulin dimers in cell lysates [33].
- Cell Lysis & Stabilization: Lyse cells or tissues in a specialized stabilization buffer containing protease inhibitors. This buffer is designed to preserve the native ratio of microtubules to free tubulin upon cell disruption. Keep samples on ice.
- Fractionation by Ultracentrifugation: Centrifuge the lysate at ultrahigh speed (100,000× g). This pellets the large, polymerized microtubules, leaving the soluble, unpolymerized tubulin in the supernatant.
- Fraction Processing: Carefully remove and retain the supernatant (free tubulin). Resuspend the pellet (microtubules) in a depolymerizing buffer.
- Quantification: Analyze both fractions by SDS-PAGE and Western blotting using an anti-tubulin antibody. The ratio of tubulin signal in the pellet to the supernatant provides the microtubule:tubulin ratio [33].
Protocol 2: In Vitro Microtubule Binding Assay (Co-sedimentation)
This assay determines if a protein binds directly to microtubules and estimates its binding affinity (Kd) [34].
- Microtubule Polymerization: Polymerize purified tubulin (4 mg/mL) in BRB80 buffer with GTP for 20 minutes at 37°C. Stabilize the resulting microtubules with taxol (final ~20 µM).
- Reaction Setup: Incubate your protein of interest with stabilized microtubules in a final volume of 50 µL (BRB80-T) for 30 minutes at room temperature.
- Separation via Cushion: Carefully layer each reaction on top of a 100 µL cushion of BRB80 buffer containing glycerol and taxol in an ultracentrifuge tube.
- Centrifugation: Pellet the microtubules and any bound protein by centrifuging at 100,000× g for 30 minutes at 23°C.
- Analysis:
- Carefully remove the supernatant (unbound protein).
- Resuspend the pellet (microtubules + bound protein) in Laemmli buffer.
- Analyze equal proportions of supernatant and pellet fractions by SDS-PAGE. Coomassie staining or immunoblotting reveals the amount of protein bound to microtubules versus unbound [34].
Experimental Workflow and Concept Visualization
Microtubule State Assay Workflow
Microtubule Lattice Dynamics and Defect Repair
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Microtubule Content and Dynamics Assays
| Reagent / Material | Function / Application | Key Characteristics |
|---|---|---|
| Microtubule/Tubulin In Vivo Assay Biochem Kit (e.g., BK038) | Measures the in vivo ratio of polymerized microtubules to free tubulin in cell/tissue lysates [33]. | Includes lysis/stabilization buffer, protease inhibitors, and tubulin standard. Semi-quantitative, uses ultracentrifugation and Western blot. |
| Paclitaxel (Taxol) | Chemotherapeutic drug that stabilizes microtubules by promoting expanded lattice spacing [3]. Used in vitro to suppress dynamics. | Nucleates and binds expanded lattices. Competes with compacting MAPs like DCX. Essential for stabilizing microtubules in binding assays [3] [34]. |
| Doublecortin (DCX) | Neuronal Microtubule-Associated Protein (MAP) that compacts microtubule lattice spacing [3]. | Used as a tool to study compaction. Its cellular localization serves as a probe for lattice state; relocalizes upon paclitaxel-induced expansion [3]. |
| Tau Protein (e.g., 2N4R isoform) | Neuronal MAP that modulates lattice dynamics, promotes bundling, and stabilizes tips [4]. | Binds diffusively along lattice. Accelerates tubulin exchange at defect sites by increasing lattice anisotropy. Challenges view of tau as purely passive stabilizer [4]. |
| GMPCPP | Non-hydrolysable GTP analog used to nucleate microtubules and create stable "caps" [35] [4]. | Stabilizes microtubule ends for studies of lattice dynamics independent of tip dynamics. Used to create seeds for growth and to cap microtubules [4]. |
| BRB80 Buffer | Standard biochemical buffer for microtubule experiments (80 mM PIPES, 1 mM MgClâ‚‚, 1 mM EGTA, pH 6.8) [34]. | Provides physiologically relevant pH and ionic conditions for tubulin polymerization and MAP interactions. |
| Reverse transcriptase-IN-1 | Reverse transcriptase-IN-1, MF:C25H17N7O2, MW:447.4 g/mol | Chemical Reagent |
| ARCC-4 | ARCC-4, MF:C53H56F3N7O7S2, MW:1024.2 g/mol | Chemical Reagent |
Modulating Lattice Stability: MAPs, Drugs, and Mechanistic Insights
FAQs: Core Concepts and Common Challenges
Q1: What is the fundamental mechanistic difference between a microtubule stabilizer and a destabilizer?
A1: Both classes ultimately suppress microtubule dynamics, but they achieve this through opposite effects on polymer mass. Microtubule stabilizers, such as paclitaxel, promote microtubule assembly and polymerization, leading to inappropriately stable microtubules that resist depolymerization [36] [37]. Conversely, microtubule destabilizers, such as vinca alkaloids, inhibit microtubule polymerization and promote depolymerization, leading to a loss of microtubule polymer mass [38] [39]. At low, clinically relevant concentrations, both classes share the net effect of suppressing microtubule dynamics without massively altering overall polymer mass, which disrupts the delicate dynamics required for proper mitotic spindle function [38] [40].
Q2: My experiment shows multipolar spindles after MTA treatment instead of mitotic arrest. Is this normal?
A2: Yes, this is a recognized and clinically relevant mechanism of action. Historically, MTAs were thought to work primarily by inducing prolonged mitotic arrest. However, recent evidence indicates that at low, therapeutically achievable concentrations, they frequently induce the formation of transient or sustained multipolar spindles [38]. Cells may then proceed through mitosis and divide on these abnormal spindles, leading to severe chromosomal instability (CIN) and cell death, often in the subsequent cell cycles [38]. This mechanism is conserved across diverse clinically useful MTAs, both stabilizers and destabilizers [38].
Q3: What are the key binding sites on tubulin for these agents?
A3: Tubulin features multiple distinct binding pockets. The classical sites are the taxane-site for stabilizers and the vinca- and colchicine-sites for destabilizers [36] [39] [40]. Research has now mapped at least nine distinct binding pockets, including emerging sites such as those for maytansine, laulimalide/peloruside A, pironetin, and gatorbulin, which offer new pharmacological entry points [36] [39] [41].
Q4: How can I overcome resistance to MTAs in my cell models?
A4: Resistance can arise from multiple mechanisms, including overexpression of drug efflux pumps like P-glycoprotein or mutations in tubulin isotypes [42]. Potential strategies to overcome resistance include:
- Combination Therapy: Combining MTAs with other agents. For example, inhibiting the p38-MK2 signaling pathway can sensitize cancer cells to MTA treatment [42].
- Novel Agents: Developing MTAs that bind to novel, emerging sites on tubulin to bypass resistance associated with classical sites [36] [39].
- Alternative Formulations: Using antibody-drug conjugates (ADCs) or nanoparticle-based delivery systems to enhance tumor-specific delivery and reduce off-target effects [36] [43].
Troubleshooting Guides
Table 1: Common Experimental Issues with MTA Treatment
| Problem Observed | Potential Cause | Suggested Solution |
|---|---|---|
| Lack of mitotic arrest phenotype | Drug concentration is too low; cells are dividing via multipolar spindles [38]. | Titrate the MTA to a low, clinically relevant nM range (e.g., 1-10 nM) and use live-cell imaging to monitor mitotic progression and spindle morphology [38]. |
| High toxicity in non-transformed control cells | Lack of selectivity; off-target effects [42]. | Optimize drug concentration. Consider trying newer agents or combinations (e.g., with a p38-MK2 inhibitor) that may show higher specificity for cancer cells [42]. |
| Variable response between cell lines | Differences in tubulin isotype expression, efflux pump activity, or intrinsic resistance mechanisms [42]. | Characterize the tubulin isotype profile (e.g., Class III β-tubulin) and P-glycoprotein status of your cell lines. Use a resistant line as a negative control. |
| Inefficient drug delivery in vivo | Poor pharmacokinetics; inability to penetrate target tissue (e.g., blood-brain barrier) [36] [41]. | Utilize advanced delivery systems such as nanoparticles, liposomes, or antibody-drug conjugates (ADCs) to improve bioavailability and targeted delivery [36] [43]. |
Table 2: Quantifying MTA Effects on Microtubule Dynamics
| Parameter | Stabilizers (e.g., Paclitaxel) | Destabilizers (e.g., Vinblastine) | Key Assays |
|---|---|---|---|
| Microtubule Polymer Mass | Increases [38] [37] | Decreases (at high doses) [38] | Tubulin polymerization assays; Western blotting for polymer vs. soluble tubulin. |
| Microtubule Dynamics | Suppresses dynamics; reduces catastrophe frequency [40]. | Suppresses dynamics; reduces growth rate [40]. | Live-cell imaging with plus-end binding proteins (e.g., EB3-GFP). |
| Mitotic Spindle Phenotype | Multipolar or grossly elongated spindles [38]. | Multipolar or fragmented spindles; loss of spindle integrity [38]. | Immunofluorescence (α/β-tubulin staining, γ-tubulin for centrosomes). |
| Cell Fate | Cell death post-mitotic exit; chromosomal instability [38]. | Cell death post-mitotic exit; chromosomal instability [38]. | Time-lapse microscopy, clonogenic assays, FACS analysis for DNA content. |
Experimental Protocols
Protocol 1: Assessing MTA Efficacy via Live-Cell Imaging of Mitotic Progression
Objective: To visualize and quantify the real-time effects of MTAs on mitotic spindle formation and cell division.
Materials:
- Cell line of interest (e.g., MDA-MB-231 breast cancer cells).
- Microtubule-Targeting Agent (MTA) stock solution (e.g., Paclitaxel, Vinblastine).
- Live-cell imaging chamber and compatible medium.
- Microscope with environmental control (37°C, 5% CO₂) and time-lapse capability.
- Fluorescent histone (e.g., H2B-GFP) to label chromosomes.
- Fluorescent tubulin marker (e.g., mCherry-α-tubulin) to visualize microtubules.
Method:
- Cell Preparation: Plate cells expressing H2B-GFP and mCherry-α-tubulin onto a live-cell imaging dish. Allow cells to adhere and grow to 40-60% confluence.
- Drug Treatment: Prepare a working solution of the MTA in pre-warmed culture medium. For initial experiments, use a low nM concentration range (e.g., 1-10 nM) to mimic clinically relevant conditions [38]. Replace the medium with the drug-containing medium.
- Image Acquisition: Immediately place the dish on the microscope stage. Acquire images every 5-10 minutes for 24-48 hours using a 40x or 60x objective. Ensure that multiple positions are captured per condition.
- Data Analysis:
- Mitotic Timing: Measure the time from nuclear envelope breakdown (NEBD) to anaphase onset or cell death.
- Spindle Morphology: Score the percentage of cells that enter mitosis with bipolar, multipolar, or monopolar spindles.
- Cell Fate Tracking: Follow individual cells to determine if they die in mitosis, exit mitosis (with or without division), or produce viable progeny.
Protocol 2: Tubulin Polymerization Assay
Objective: To biochemically determine whether a compound acts as a stabilizer or destabilizer by measuring its effect on microtubule polymer mass.
Materials:
- Purified tubulin proteins.
- Test compound (MTA).
- GTP and polymerization buffer (e.g., PEM: PIPES, EGTA, MgClâ‚‚).
- Pre-warmed microcentrifuge tubes and a thermosafe centrifuge.
- SDS-PAGE equipment.
Method:
- Reaction Setup: Prepare a solution of purified tubulin (e.g., 2 mg/mL) in polymerization buffer containing GTP. Aliquot the solution into separate tubes.
- Treatment: Add the test MTA to the experimental tubes. Include a vehicle control (e.g., DMSO) and a known stabilizer (e.g., paclitaxel) and destabilizer (e.g., nocodazole) as controls.
- Polymerization: Incubate the reactions at 37°C for 30-60 minutes to allow microtubule polymerization.
- Separation: Centrifuge the samples at high speed (e.g., 100,000 x g) at 37°C to pellet polymerized microtubules. Carefully separate the supernatant (soluble tubulin) from the pellet (polymerized tubulin).
- Analysis: Resuspend the pellet in a known volume of buffer. Analyze both the supernatant and pellet fractions by SDS-PAGE. Quantify the amount of tubulin in each fraction via Western blot or Coomassie staining. A stabilizer will increase tubulin in the pellet fraction, while a destabilizer will increase it in the supernatant [38].
Pathway and Mechanism Visualization
Microtubule Dynamics and MTA Mechanisms
MTA-Induced Cell Fate Decision Pathway
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for MTA Research
| Reagent | Function & Application | Key Considerations |
|---|---|---|
| Paclitaxel (Taxol) | Classical microtubule stabilizer. Used to study mitotic arrest, spindle assembly, and chromosomal instability [38] [37]. | At high doses (>1 µM) induces mitotic arrest; at low nM doses induces multipolar spindles and CIN [38]. |
| Vinblastine / Vinorelbine | Classical microtubule destabilizers. Used to inhibit polymerization and study consequences of microtubule depolymerization [38] [39]. | High concentrations depolymerize microtubules; low concentrations suppress dynamics without affecting mass [38]. |
| [¹¹C]MPC-6827 | A positron emission tomography (PET) radiotracer that selectively binds to destabilized microtubules. Enables in vivo visualization of MT dynamics in neurodegenerative models and potentially oncology [44]. | Emerging tool for translational research; allows non-invasive monitoring of microtubule stability. |
| EB3-GFP (or similar +TIP protein) | A plus-end tracking protein. Used in live-cell imaging to visualize and quantify microtubule dynamics (growth speed, catastrophe frequency) in real-time [44] [40]. | Critical for assessing the subtle effects of low-dose MTAs on dynamics rather than bulk polymer mass. |
| CMPD1 | A dual-target inhibitor that suppresses the p38-MK2 pathway and acts as a microtubule destabilizer. Useful for combination studies to sensitize cells to other MTAs [42]. | Demonstrates the potential of hybrid or multi-target agents in overcoming resistance. |
| Antibody-Drug Conjugates (ADCs) e.g., Trastuzumab-emtansine | Targeted delivery system consisting of a monoclonal antibody linked to a cytotoxic MTA payload (e.g., maytansinoid). Used to improve tumor specificity and reduce off-target toxicity [43]. | Key for translational research, modeling targeted therapy efficacy and resistance mechanisms. |
| PROTAC EED degrader-2 | PROTAC EED degrader-2 is a potent, VHL-based degrader of the PRC2 complex (EED, EZH2, SUZ12). For research use only. Not for human or veterinary use. |
Frequently Asked Questions (FAQs)
Q1: What is the fundamental paradox in Tau's function on microtubules? The paradox is that Tau simultaneously accelerates the exchange of individual tubulin dimers within the microtubule lattice while also slowing the overall fracture and depolymerization of the microtubule. Traditionally, Tau was viewed as a simple stabilizer that reduces microtubule dynamics. Recent research shows it is an active remodeler that enhances lattice dynamics (specifically, tubulin turnover) while simultaneously improving the microtubule's structural resilience [4] [45].
Q2: How does Tau accelerate tubulin exchange without causing microtubule disintegration? Tau promotes tubulin exchange predominantly at pre-existing topological defect sites in the lattice. At these sites, it alters the energy landscape of tubulin-tubulin interactions by stabilizing longitudinal bonds between dimers along the protofilament, while destabilizing lateral bonds between adjacent protofilaments [4]. This "lattice anisotropy" facilitates the repair of defects. The exchange is localized and coupled with a self-repair process, leading to a net stabilization effect [4].
Q3: What is the molecular mechanism behind Tau's dual role? The mechanism involves Tau's binding at the interface between tubulin heterodimers. Biochemical and NMR studies indicate that Tau binds to a hydrophobic pocket on α-tubulin that overlaps with the vinblastine binding site, a site crucial for regulating microtubule polymerization [46]. This interaction allows Tau to allosterically influence the stability of the tubulin lattice, promoting a conformation that favors longitudinal stability and facilitates lateral tubulin incorporation and loss [4] [46].
Q4: How does phosphorylation affect Tau's function in microtubule binding? Phosphorylation, particularly in the proline-rich region of Tau, acts as a graded regulatory mechanism. The overall number of phosphorylated sites, rather than the specific locations, dominantly modulates Tau's affinity for the microtubule lattice [47]. Increased phosphorylation reduces microtubule binding, which can tune Tau's role in tubulin exchange and fracture resistance. In pathological hyperphosphorylation, this can lead to a significant loss of Tau's physiological functions [47] [48].
Troubleshooting Guides
Problem: Inconsistent Results in Tubulin Incorporation Assays
| Potential Cause | Solution |
|---|---|
| Uncontrolled Tau Concentration | Maintain low, physiologically relevant Tau concentrations (e.g., 0.5-20 nM). High concentrations can lead to overcrowding and obscure the acceleration effect [4]. |
| Insufficient Incubation Time | Extend the incubation time with labelled tubulin to 15-30 minutes to clearly observe tau-enhanced incorporation stretches [4]. |
| Microtubule Lattice Defect Variability | Use consistent methods for preparing capped microtubules. Be aware that the number of inherent lattice defects can vary between preparations and affect the baseline rate of tubulin exchange [4]. |
Problem: Failure to Observe Tau-Induced Slowing of Microtubule Fracture
| Potential Cause | Solution |
|---|---|
| Presence of Free Tubulin | Ensure the fracture assay buffer is free of tubulin to prevent lattice repair from solution, which would mask the fracture-slowing effect of Tau [4]. |
| Microtubules Not Properly Capped | Verify the efficiency of the capping step with GMPCPP-tubulin. Uncapped dynamic ends will depolymerize rapidly, overwhelming the lattice stabilization effect [4]. |
| Tau Isoform or Phosphorylation State | Use a well-characterized, recombinant Tau isoform (e.g., human 2N4R). Check if phosphorylation (e.g., from baculovirus expression) reduces its activity; use bacterially expressed Tau for non-phosphorylated experiments [49]. |
Table 1: Key Quantitative Findings from Tau Lattice Dynamics Studies
| Parameter | Condition 1 (0 nM Tau) | Condition 2 (20 nM Tau) | Measurement Method | Source |
|---|---|---|---|---|
| Median Tubulin Incorporation Length | 0.7 µm | 1.2 µm | Tubulin incorporation assay | [4] |
| Median Distance Between Incorporation Sites | 12.4 µm | 6.6 µm | Tubulin incorporation assay | [4] |
| Overall Tubulin Incorporation | 1x (Baseline) | 4x increase | Tubulin incorporation assay | [4] |
| Lattice Fluorescence Reduction at Incorporation Sites | ~7-15% (1-2 protofilaments) | ~7-15% (1-2 protofilaments) | Fluorescence intensity quantification | [4] |
| Tau Residence Time on Microtubule | - | ~3.7 seconds | Fluorescence Recovery After Photobleaching (FRAP) | [4] |
| Tau Diffusion Coefficient on Lattice | - | ~0.2 µm²/s | Single-particle tracking / FRAP | [4] |
Table 2: Research Reagent Solutions for Key Experiments
| Reagent / Material | Function in Experiment | Key Details / Considerations |
|---|---|---|
| Human 2N4R Tau | The primary MAP under study. | The longest isoform; commonly used to study full functionality. Ensure expression system is noted (bacterial vs. baculovirus) [49]. |
| GMPCPP-tubulin | Forms stable "caps" on microtubule ends. | Inhibits microtubule tip dynamics, allowing isolated study of lattice dynamics [4]. |
| GTP-tubulin (fluorescently labelled) | Visualizes tubulin incorporation into the lattice. | Use low concentrations (e.g., 8 µM) to minimize tip growth during incorporation steps [4]. |
| Primary Cultured Rat Hippocampal Neurons | For studying Tau dynamics in a physiological cellular context. | Used in FRAP experiments to analyze microtubule-binding dynamics [47]. |
| Vinblastine | A competitive inhibitor for Tau binding. | Used to map Tau's binding site to the vinblastine pocket on α-tubulin [46]. |
Experimental Protocols
Protocol 1: Measuring Tau-Accelerated Tubulin Exchange
Objective: To visualize and quantify the effect of Tau on the incorporation of new tubulin dimers into the existing microtubule lattice.
Methodology:
- Seed Preparation: Grow dynamic microtubules from surface-attached, stable GMPCPP-microtubule seeds using green-labelled GTP-tubulin [4].
- Capping: Inhibit further tip dynamics by capping the microtubules with a second layer of GMPCPP-tubulin [4].
- Incorporation Incubation: Incubate the capped microtubules with red-labelled GTP-tubulin (e.g., 8 µM) in the presence of your experimental Tau concentration (e.g., 0, 0.5, 20 nM) for 15-30 minutes [4].
- Wash and Image: Wash out free tubulin to reduce background and image the microtubules to visualize red-labelled incorporation stretches [4].
- Quantification: Measure the length of incorporation stretches, the distance between them, and the overall fraction of the lattice that has incorporated new tubulin [4].
Protocol 2: Microtubule Fracture Assay to Probe Stabilization
Objective: To assess the protective effect of Tau against microtubule breakage under mechanical stress or intrinsic lattice weakness.
Methodology:
- Grow and Cap Microtubules: Follow steps 1 and 2 from Protocol 1 to create capped microtubules [4].
- Induce Fracture: Image the microtubules in a tubulin-free buffer in the presence or absence of Tau (e.g., 20 nM). The absence of free tubulin promotes net tubulin loss from the lattice, leading to fracture over time [4].
- Image and Analyze: Record time-lapse images to capture fracture events. Quantify the time-to-fracture or the rate of fracture for microtubules under different conditions [4].
Conceptual Diagrams
Tau's Dual Role Mechanism
Experimental Workflow: Tubulin Exchange Assay
How Lattice Anisotropy Influences Damage Propagation and Repair
Frequently Asked Questions (FAQs)
FAQ 1: What is microtubule lattice anisotropy and why is it critical for stability?
Microtubule lattice anisotropy refers to the directional dependence of bonding energies between tubulin dimers within the microtubule lattice. It is defined as the ratio of longitudinal to lateral binding energies (A = ΔG_long / ΔG_lat). This anisotropy determines how fractures propagate; higher values favor longitudinal crack growth, while lower values promote lateral spreading. It is fundamental to the lattice's mechanical strength and its ability to contain damage [10].
FAQ 2: How does the tau protein actively repair microtubule lattice defects?
Contrary to being a passive stabilizer, tau acts as an active repair catalyst. It increases lattice anisotropy by stabilizing longitudinal tubulin-tubulin interactions while simultaneously destabilizing lateral ones. This change in energy landscape enhances the mobility of lattice defects, promoting their annihilation and facilitating the incorporation of new tubulin, thereby enabling efficient self-repair without increasing the net rate of tubulin loss [4].
FAQ 3: What role do 'seams' play in damage propagation?
Seams are structural discontinuities in the microtubule lattice that act as pre-existing pathways for accelerated damage propagation. When a growing crack front encounters a seam, the dimer at the boundary gains an additional lateral contact, which can significantly increase the longitudinal propagation speed of the fracture. Multiple seams emanating from a single defect can break propagation symmetry, leading to faster fracture in one direction [10].
FAQ 4: What are the key experimental parameters for quantifying lattice dynamics?
Critical parameters include the binding energies for longitudinal (ΔG_long) and lateral (ΔG_lat) contacts, the lattice anisotropy ratio (A), and the off-rate constant of a corner dimer (1/τ). These parameters allow researchers to model detachment rates for dimers with varying numbers of neighbors using an Arrhenius equation framework, which is essential for predicting fracture behavior [10].
Troubleshooting Guides
Problem: Inconsistent Microtubule Fracture Times in Experiments
Issue: Experimentally observed fracture times deviate significantly from model predictions.
Solution:
- Check Defect Type: Confirm the initial defect structure. Monomer vacancies, especially those adjacent to seams, drastically reduce fracture time compared to dimer vacancies [10].
- Verify Lattice Anisotropy: Re-evaluate the assumed ratio of longitudinal to lateral binding energies (
A). Recent studies suggest the intrinsicAvalue is bounded at approximately 1.5, challenging earlier, higher predictions [10]. - Account for Seams: Ensure your model incorporates the accelerating effect of seams on fracture propagation. A defect's position relative to a seam is a critical factor [10].
Problem: Low Tubulin Incorporation in Lattice Repair Assays
Issue: Inability to observe significant tubulin exchange within the microtubule lattice.
Solution:
- Introduce Tau Protein: Supplement with low concentrations (e.g., 20 nM) of human 2N4R tau. Tau can accelerate tubulin exchange at defect sites, leading to longer incorporation stretches and a higher spatial frequency of repair events [4].
- Ensure Defect Presence: Remember that tubulin exchange occurs predominantly at pre-existing topological defect sites. Assays using perfectly stable, defect-free lattices will show minimal activity [4].
- Optimize Incubation Time: Extend the incubation time with labelled tubulin. In the presence of tau, incorporation stretches can cover a significant portion of the lattice after 30 minutes [4].
Table 1: Key Parameters from Microtubule Lattice Fracture Models
| Parameter | Description | Value / Range | Significance |
|---|---|---|---|
| A (Anisotropy) | Ratio of longitudinal to lateral binding energy (ΔG_long / ΔG_lat) |
~1.5 (bound) [10] | Determines the directionality of fracture propagation. |
| Fracture Time | Time for a damage zone to span all protofilaments. | 10 - 20 minutes [10] | Key observable for validating kinetic models against experiment. |
| Damage Zone Size | Average length of damaged lattice before fracture. | ~1 µm [10] | Relates to the critical defect size for catastrophic failure. |
| v_long^seam | Longitudinal fracture propagation speed at a seam. | ≥ η kâ‚₃ (Accelerated) [10] | Seams act as pre-existing pathways that accelerate damage. |
Table 2: Experimental Parameters for Tau-Induced Lattice Repair
| Experimental Condition | Incorporation Length (median) | Distance Between Incorporations (median) | Key Finding |
|---|---|---|---|
| No Tau (15 min) | 0.7 µm | 12.4 µm | Baseline lattice dynamics. |
| 20 nM Tau (15 min) | 1.2 µm | 6.6 µm | Tau significantly enhances longitudinal incorporation and event frequency. |
| 20 nM Tau (30 min) | Extensive (up to full lattice) | No further increase | Saturation of incorporation sites or event overlap. |
Detailed Experimental Protocols
Protocol 1: Kinetic Monte Carlo Simulation of Microtubule Fracture
This protocol is based on the model used to study fracture pathways from initial defects [10].
- System Representation: Model the microtubule as a two-dimensional lattice at the monomer scale. For a standard 13-protofilament MT, this is a grid of 13 (lateral) by N (longitudinal) positions.
- Define Lattice Parameters:
- Set the longitudinal (
ΔG_long) and lateral (ΔG_lat) binding energies. - Calculate the lattice anisotropy
A = ΔG_long / ΔG_lat. - Set the off-rate constant for a corner dimer (
1/Ï„).
- Set the longitudinal (
- Initialize Defect: Introduce an initial defect into the lattice, such as a monomer vacancy. Specify its location (e.g., in the B-lattice, or adjacent to a seam).
- Implement Kinetic Rules: The detachment rate
k_mnfor a dimer withmlongitudinal andnlateral neighbors is governed by the Arrhenius equation:k_mn = (1/τ) * exp( β * ( m * ΔG_long + (n/2) * ΔG_lat - ΔG_b/2 ) )whereβis the inverse thermodynamic temperature andΔG_bis the total binding energy of a fully surrounded dimer. - Run Simulation: Using a kinetic Monte Carlo algorithm, simulate the stochastic detachment of dimers over time. Track the growth of the vacancy until it spans all 13 protofilaments, marking complete fracture.
- Analysis: Quantify the time to fracture and the shape of the damage zone under different initial conditions and anisotropy values.
Protocol 2: Measuring Tau-Mediated Tubulin Exchange In Vitro
This protocol outlines the method for visualizing tau's effect on lattice dynamics [4].
- Microtubule Preparation:
- Step I: Grow dynamic microtubules from surface-attached, GMPCPP-stabilized seeds using green-fluorescently labelled GTP-tubulin.
- Step II: Cap the microtubule ends with GMPCPP-tubulin to inhibit further tip dynamics.
- Tubulin Incorporation Phase:
- Step III: Incubate the capped microtubules with red-fluorescently labelled GTP-tubulin (e.g., 8 µM) in the presence of defined concentrations of tau (e.g., 0 nM, 0.5 nM, 20 nM) for a set duration (e.g., 15-30 minutes).
- Visualization:
- Step IV: Wash out free tubulin and tau to reduce background fluorescence.
- Image the microtubules using TIRF or other high-resolution fluorescence microscopy.
- Data Analysis:
- Quantify the length and spatial frequency of red tubulin incorporation stretches.
- Measure the fluorescence intensity of the original (green) lattice at incorporation sites to quantify tubulin exchange (loss).
- Compare these metrics between tau-positive and tau-negative conditions.
Experimental Workflows and Pathway Diagrams
Diagram 1: Tau-Mediated Lattice Repair Pathway (Width: 760px)
Diagram 2: Tubulin Exchange Assay Workflow (Width: 760px)
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Microtubule Lattice Studies
| Item | Function / Description | Application Example |
|---|---|---|
| GMPCPP-tubulin | Slowly hydrolysable GTP analogue that stabilizes microtubule ends and seeds. | Used to create stable seeds for growth and to cap dynamic ends in lattice exchange assays [4]. |
| Fluorophore-labelled GTP-tubulin (e.g., Green, Red) | Tubulin conjugated to fluorescent dyes for visualization. | Allows tracking of microtubule growth, lattice incorporation, and turnover via fluorescence microscopy [4]. |
| Recombinant Tau Protein (e.g., human 2N4R isoform) | A key microtubule-associated protein (MAP) that modulates lattice dynamics. | Used to investigate the active role of MAPs in promoting lattice repair and altering anisotropy [4]. |
| Kinetic Monte Carlo Simulation Code | Custom software for simulating dimer-level lattice dynamics. | Employed to model fracture propagation from defects and deduce critical lattice energy parameters [10]. |
| All-Atom Molecular Dynamics (MD) | High-resolution computational modeling of molecular interactions. | Used to simulate detailed interactions at microtubule tips and validate coarse-grained models [50]. |
Technical Support Center: Troubleshooting Guides and FAQs
This section addresses common experimental challenges in drug characterization, with a specific focus on assays relevant to cytoskeletal research, such as microtubule dynamics.
Frequently Asked Questions (FAQs)
FAQ 1: Our cell-based high-throughput screening (HTS) assay shows high variability in viability readouts. How can we improve its robustness?
A robust and reproducible cell-based assay is essential for reliable high-throughput screening. Key optimization variables must be carefully controlled to minimize variability and maximize the signal-to-noise ratio [51].
- Assay Performance Metrics: Calculate the Z'-factor to statistically assess the quality and robustness of your assay. A Z'-factor ≥ 0.5 is generally indicative of an excellent assay suitable for HTS, as it reflects a wide separation between positive and negative controls [51].
- Critical Optimization Steps:
- Cell Seeding Density: Titrate the number of cells per well to find the optimal density that avoids both overcrowding and under-representation, ensuring a linear response [51].
- Incubation Time: Determine the optimal duration for drug exposure before measuring viability (e.g., 24, 48, 72 hours) [51].
- Reagent Concentration: Titrate the concentrations of dyes or substrates to achieve the best signal-to-noise ratio while ensuring the reagents themselves are not toxic to the cells [51].
- Control and Normalization: Always include a positive control (e.g., Staurosporine for cytotoxicity) and a negative control (e.g., DMSO vehicle) on every plate. Normalize all results to these controls to account for plate-to-plate variability [51].
FAQ 2: What are the key considerations for transitioning from a biochemical to a more physiologically relevant cellular assay for target engagement?
Moving to a cellular context is critical for confirming pharmacological activity within a biological system. This is especially important for complex targets like microtubules, where the cellular environment significantly influences drug action [52] [3].
- Confirm Target Engagement in Cells: Utilize methods that validate direct binding in intact cells. The Cellular Thermal Shift Assay (CETSA) has emerged as a leading approach. It can quantitatively confirm dose-dependent and target-specific engagement, closing the gap between biochemical potency and cellular efficacy [52].
- Embrace Complex Cell Models: For ADME (Absorption, Distribution, Metabolism, and Excretion) and toxicity questions, simple 2D cultures have limitations. Consider adopting more complex cell models such as 3D spheroids, co-cultures, or organ-on-a-chip systems. These models better replicate the tissue-specific mechanical and biochemical characteristics of target organs, providing more predictive data for human outcomes [53] [54].
- Leverage Computational Tools Early: Integrate PBPK (Physiologically Based Pharmacokinetic) modelling and simulation during the discovery phase. This can help bridge drug discovery and development by predicting human pharmacokinetics, oral absorption, and potential drug-drug interactions, thereby increasing the chances of a candidate's success [54].
FAQ 3: Our research involves characterizing compounds that affect microtubule dynamics. What in vitro and cellular tools can we use to characterize their mechanism?
Characterizing the impact of compounds on a dynamic cellular structure like the microtubule cytoskeleton requires a multi-faceted approach.
- In Vitro Reconstitution: Employ in vitro buckling assays to directly observe the effects of a compound on microtubule lattice spacing using light microscopy. This assay can demonstrate whether a molecule, like paclitaxel, acts as a microtubule "expander" or if another protein, like DCX, acts as a "compactor" [3].
- Cellular Probing with +TIPs: In cells, use microtubule plus-end tracking proteins (+TIPs) as biosensors. The localization and behavior of +TIPs (e.g., CLASP, APC, or EB1) are highly sensitive to changes in microtubule dynamics and stability downstream of signaling cues. For example, the relocalization of DCX in response to paclitaxel treatment serves as a probe for drug-induced microtubule expansion [55] [3].
- High-Content Screening (HCS): Implement High-Content Screening or Cell Painting assays. These combine high-resolution microscopy, fluorescent labeling (e.g., of tubulin), and sophisticated image analysis to capture complex cellular phenotypes, such as changes in microtubule organization, organelle structure, and cell morphology [51].
Troubleshooting Common Experimental Issues
Issue 1: Inconsistent Results in In Vitro Transporter Interaction Assays
- Problem: Data from transporter interaction studies (e.g., for Drug-Drug Interaction or DDI assessments) is not reproducible.
- Solution:
- Follow Harmonized Guidelines: Adhere to the ICH M12 guideline, which aims to harmonize international regulatory guidance on DDI studies. This ensures your assay design and data interpretation meet global standards [54].
- Assay Advancements: Utilize improved methods for challenging compounds, such as novel techniques for the accurate measurement of plasma protein binding for highly bound compounds, which can significantly impact results [54].
Issue 2: Poor Translational Predictivity from Animal Models to Human Clinical Trials
- Problem: Drug candidates show promising results in animal models but fail due to toxicity or lack of efficacy in humans.
- Solution:
- Adopt Advanced In Vitro Models: Integrate microphysiological systems (MPS), such as gut-liver-on-a-chip models. These systems more closely mimic human physiology and can better predict human-specific adverse effects like Drug-Induced Liver Injury (DILI), addressing the significant limitations of traditional animal models [53].
- Incorporate Human-Relevant Cells: Use induced pluripotent stem cell (iPSC)-derived hepatocytes or other cell types within these advanced platforms to create a more human-relevant system for preclinical safety assessment [53].
Quantitative Data for Experimental Design
This section provides consolidated quantitative data to inform the design and troubleshooting of characterization assays.
Table 1: Key Metrics for Robust Cell-Based Assay Development
| Parameter | Optimal Value or Method | Technical Note |
|---|---|---|
| Assay Quality (Z'-factor) | ≥ 0.5 | Benchmark for an excellent HTS assay; indicates a wide signal window [51]. |
| Cell Seeding | Titrated density | Avoid overcrowding/under-representation; ensure linear response [51]. |
| Control Selection | Positive (e.g., Staurosporine) & Negative (e.g., DMSO) | Essential for defining maximal response and baseline on every plate [51]. |
| Incubation Time | Variable (e.g., 24, 48, 72 hrs) | Must be optimized for the specific drug and cell model [51]. |
| Readout Methods | ATP-based (luminescence), Resazurin (fluorescence), MTT (colorimetric) | Selection depends on required sensitivity, compatibility, and instrumentation [51]. |
Table 2: Microtubule Lattice Spacing States and Regulatory Factors
| State / Factor | Lattice Spacing (Ã…) | Description & Regulatory Influence |
|---|---|---|
| Expanded State | 83.5 ± 0.2 | GTP-like lattice; promoted by kinesin-1 and the chemotherapeutic drug paclitaxel [3]. |
| Compacted State | 81.7 ± 0.1 | GDP-like lattice; promoted by the neuronal MAP doublecortin (DCX) [3]. |
| MAPs as Compactors | N/A | Proteins like doublecortin (DCX) and EB-family proteins nucleate compacted lattices or bind preferentially to them [3]. |
| MAPs as Expanders | N/A | Proteins like kinesin-1 can directly expand pre-formed GDP lattices, impacting intracellular trafficking [3]. |
Experimental Protocols for Key Assays
Protocol 1: High-Throughput Cell Viability Screening
This protocol outlines a standardized process for screening compound libraries using cell-based viability assays [51].
Step-by-Step Workflow:
- Plating Cells: Use automated liquid handlers to uniformly dispense cells into multi-well plates (96-, 384-, or 1536-well). Incubate plates under standard conditions (37°C, 5% CO₂) to allow cell adhesion.
- Compound Addition: Employ robotic systems to transfer precise volumes of individual compounds from library source plates to the assay plates. Include positive and negative controls on each plate.
- Viability Measurement: After an appropriate drug incubation period, add a homogeneous viability reagent (e.g., an ATP-based luminescent assay) directly to the wells.
- Detection & Analysis: Use an integrated microplate reader to detect the signal (luminescence/fluorescence/absorbance). Analyze the data with specialized software, normalizing to controls to identify "hit" compounds.
Protocol 2: Characterizing Compound Effects on Microtubule Lattice Spacing
This protocol describes a methodology for investigating whether a compound influences microtubule lattice structure, using a combination of in vitro and cellular techniques [3].
Step-by-Step Workflow:
- In Vitro Buckling Assay:
- Create "double-capped" microtubules with a long GDP-lattice segment stabilized between two GMPCPP caps.
- Introduce the compound of interest and observe the microtubules under light microscopy.
- Interpretation: Compound-induced lattice expansion will manifest as an increase in the length of the GDP-segment, causing the microtubule to buckle under axial compression.
- Cellular Localization Assay:
- Transfert cells to express a fluorescently tagged microtubule-associated protein (MAP) known to be sensitive to lattice spacing, such as doublecortin (DCX).
- Treat the cells with the compound and use live-cell or fixed-cell microscopy to observe the localization pattern of the MAP.
- Interpretation: Altered localization patterns (e.g., DCX relocalizing to curved segments) serve as a cellular proxy for compound-induced changes in underlying microtubule lattice structure.
Signaling Pathways and Experimental Workflows
Diagram 1: Signaling to Lattice Dynamics
Diagram 2: HTS Screening Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Tools for Drug Characterization & Microtubule Research
| Tool / Reagent | Function / Application | Example Use-Case |
|---|---|---|
| CETSA (Cellular Thermal Shift Assay) | Validates direct target engagement of drugs in intact cells and tissues [52]. | Confirming that a compound binds to its intended target (e.g., tubulin) within a cellular environment, bridging biochemical and cellular efficacy [52]. |
| +TIPs (e.g., CLASP, APC, EB1) | Endogenous biosensors for microtubule dynamics; their localization reports on microtubule stability and guidance cue signaling [55]. | Monitoring changes in microtubule behavior in response to extracellular signals or compound treatment by imaging GFP-tagged +TIPs [55]. |
| Organ-on-a-Chip (OOC) | Advanced in vitro model that mimics human organ physiology (e.g., gut-liver) for predictive ADME and toxicity testing [53]. | Assessing human-specific Drug-Induced Liver Injury (DILI) and gut-liver axis interactions of new drug candidates, reducing reliance on non-predictive animal models [53]. |
| Paclitaxel & DCX | Pharmacological tools to manipulate microtubule lattice spacing; Paclitaxel is an "expander," DCX is a "compactor" [3]. | Used in in vitro buckling assays or as cellular probes to study the impact of lattice spacing changes on microtubule function and MAP binding [3]. |
| ICH M12 Guideline | International harmonized guideline for the design and interpretation of drug-drug interaction (DDI) studies [54]. | Ensuring that in vitro transporter and metabolic DDI assessments are conducted to globally accepted regulatory standards during drug development [54]. |
Frequently Asked Questions (FAQs)
Q1: What are the primary factors that limit my ability to resolve individual microtubules within bundles? The key factor is the effective label size, which determines the apparent diameter of the microtubule in super-resolution images. Conventional primary and secondary antibody combinations can displace the fluorescent probe by 12.5 nm or more from the microtubule lattice, increasing the apparent diameter and causing neighboring microtubules to blend together. For bundled microtubules with a center-to-center spacing of 50-70 nm, this often makes resolution of individual filaments impossible [56].
Q2: How does the nucleotide state of tubulin affect my imaging experiments? The nucleotide state (GTP-bound vs. GDP-bound) controls the conformational state of the tubulin dimer within the microtubule lattice. GTP-tubulin adopts an "expanded" conformation, while hydrolysis to GDP triggers lattice "compaction." This conformational change not only regulates microtubule dynamics but also controls the accessibility of key regions, such as the C-terminal tails, for binding by probes, motor proteins, and enzymes involved in post-translational modifications [57]. This means the same structure may be more or less visible depending on its biochemical state.
Q3: My live-cell lattice light-sheet imaging is causing photodamage to sensitive samples. How can I mitigate this? Lattice light-sheet microscopy (LLSM) is specifically designed to minimize photodamage through selective plane illumination. To optimize viability [58]:
- Ensure your imaging medium is properly formulated and equilibrated. For example, for post-implantation mouse embryos, use a mix of CMRL, KnockOut serum, and L-Glutamine, and pre-equilibrate in a 37°C, 5% CO₂ incubator for at least 1 hour.
- Carefully control mounting. Use finely pulled glass capillaries to secure embryos with minimal physical stress.
- Utilize the speed of LLSM to lower the required light dose per time point, reducing cumulative phototoxicity during time-lapse experiments [58].
Q4: What are the trade-offs between different super-resolution techniques for imaging microtubule lattices? The choice of technique depends on your specific requirements for resolution, speed, and sample compatibility. The table below summarizes key characteristics:
Table 1: Comparison of Super-Resolution Techniques for Microtubule Imaging
| Technique | Theoretical Resolution (Lateral) | Key Advantages | Key Limitations / Challenges |
|---|---|---|---|
| Structured Illumination Microscopy (SIM) | ~144 nm (with deconvolution) [59] | Fast acquisition, wide field of view, lower phototoxicity [60] | Limited resolution gain; scattering disrupts patterns in deep tissue [59] |
| Single-Molecule Localization Microscopy (SMLM) | < 30 nm (dependent on label) [56] | Very high resolution | Small label size critical; requires high labeling density; background fluorescence hinders deep-tissue imaging [59] [56] |
| Lattice Light-Sheet Microscopy (LLSM) | High spatial-temporal resolution [58] | Minimal photodamage, fast 4D imaging of dynamic processes | Specialized setup; sample mounting can be complex [58] |
| Image Scanning Microscopy (ISM) | 144 nm (lateral), 351 nm (axial) [59] | Excellent optical sectioning, good balance of resolution and depth | --- |
| Stimulated Emission Depletion (STED) | Nanoscale | High resolution | Resolution loss in deep tissue due to distortion of depletion beam [59] |
Q5: Why might my biosensors or labels show unexpectedly low binding to microtubules in live cells? This is likely due to the conformational state of the microtubule lattice. Key regions, such as the C-terminal tail of α-tubulin, are not always freely accessible as traditionally assumed. Their exposure is regulated by the lattice conformation. Accessibility can be increased by [57]:
- Treating with Taxol, which stabilizes an expanded, GTP-like lattice.
- Expressing MAPs like kinesin-1 or CAMSAP3 that recognize or induce an expanded lattice.
- Using GTP-locked tubulin mutants that maintain the expanded conformation.
Troubleshooting Guide: Common Artifacts and Solutions
Table 2: Troubleshooting Common Lattice Imaging Artifacts
| Problem | Potential Cause | Solution | Supporting Experimental Protocol |
|---|---|---|---|
| Unresolvable Microtubule Bundles | Large effective label size causing blended signals [56]. | Use smaller labels such as anti-tubulin nanobodies (e.g., VHH#1/VHH#2) or directly conjugated Fab fragments. | Protocol: Testing Labels with In Vitro Bundling Assay1. Polymerize microtubules in vitro.2. Add microtubule bundling protein (e.g., AtMAP65-1) to form arrays with known ~65 nm spacing.3. Attach bundles to silanized coverslips.4. Label with your probe (e.g., nanobody-AF647).5. Perform SMLM. If individual microtubules are resolved, the label is suitable for cellular use [56]. |
| Low Signal-to-Noise in Deep Tissue | Scattering of excitation light and out-of-focus fluorescence causing background [59]. | Implement physical background suppression. Use a spinning-disk (SD) confocal unit integrated into your microscope to physically eliminate out-of-focus signals before detection [59]. | Protocol: Confocal² Spinning-Disk ISM (C2SD-ISM)1. Hardware: Integrate an SD unit and a DMD for multifocal illumination into your light path. The SD pinhole array and DMD should be conjugate to the sample plane.2. Imaging: Use the DMD to project a sparse, shifting multifocal pattern onto the sample.3. Detection: Fluorescence is filtered by the SD pinholes before reaching the sCMOS camera.4. Reconstruction: Apply a dynamic pinhole array pixel reassignment (DPA-PR) algorithm to reconstruct the super-resolution image [59]. |
| Inaccessible Epitopes on Microtubule Lattice | The target region (e.g., α-tubulin C-terminal tail) is obscured due to a compacted GDP-lattice conformation [57]. | Modulate the lattice to an expanded state. Treat cells with Taxol (e.g., 10 µM for 30 min) or express MAPs that promote expansion (e.g., kinesin-1, CAMSAP3) to increase probe accessibility [57]. | Protocol: Probing Lattice Conformation with Biosensors1. Transfert cells with plasmids for Y-αCTT biosensors (e.g., rMAb-YL1/2-GFP).2. Image live cells to establish baseline binding.3. Add lattice-modifying agent (e.g., Taxol).4. Monitor increases in microtubule binding of the biosensor in real-time as an indicator of lattice expansion and epitope exposure [57]. |
| Artifacts from Fixation | Chemical fixation can alter microtubule structure and antigen accessibility [57]. | Where possible, use live-cell imaging. If fixation is necessary, validate findings with live-cell biosensors and try different fixatives (e.g., paraformaldehyde vs. methanol) to find the one that best preserves your structure of interest. | Control Experiment: Compare the binding pattern of your probe in gently fixed cells (e.g., with 4% PFA for 10 min) versus its behavior in live cells expressing a GFP-tagged version of the same probe. A significant difference indicates fixation-induced artifacts [57]. |
Essential Workflow and Concept Diagrams
Diagram 1: Lattice Conformation Controls Probe Accessibility
Diagram 2: Super-Resolution Imaging Workflow
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Advanced Microtubule Lattice Imaging
| Reagent / Tool | Function / Description | Key Application | Considerations |
|---|---|---|---|
| Anti-Tubulin Nanobodies (VHHs) [56] | Single-domain antibody fragments (~15 kDa, ~4 nm) that bind directly to tubulin. | Super-resolution imaging (e.g., SMLM) of tightly bundled microtubules. Reduces apparent microtubule diameter to ~39 nm, enabling resolution of filaments ~50 nm apart [56]. | Must be purified and conjugated to photo-switchable dyes (e.g., AF647). Specificity for tubulin isotypes should be verified. |
| GMPCPP [61] | A non-hydrolysable GTP analog. | Stabilizes microtubules in a GTP-like, expanded conformation. Used to study the structure of stable microtubule ends and to probe nucleotide-dependent conformational changes [61] [57]. | Cost can be prohibitive for large-scale experiments. |
| Taxol/Paclitaxel [61] [57] | A natural product that stabilizes microtubules. | Allosterically induces a GMPCPP-like, expanded lattice conformation. Used to study expanded lattice states and increase accessibility of obscured epitopes like the α-tubulin C-terminal tail [61] [57]. | Its effect on lattice conformation is a crucial consideration beyond mere stabilization. |
| Tubulin Biosensors (e.g., rMAb-YL1/2) [57] | Recombinant monoclonal antibodies or other probes targeting specific tubulin epitopes (e.g., tyrosinated α-tubulin tail). | Live-cell reporting of microtubule lattice conformation and epitope accessibility. Binding indicates an expanded lattice state [57]. | Low binding in live cells under normal conditions is expected; increased binding reports on conformational change. |
| Specialized Culture Media (e.g., for embryos) [58] | Chemically defined media optimized for specific sample types. | Supports viability during long-term, high-resolution live imaging of sensitive samples like post-implantation embryos [58]. | Formulation is critical; for example, L-Glutamine crystals can cause imaging artifacts. Must be prepared fresh and properly equilibrated. |
Translational Validation: From Bench to Biomarker and Therapeutics
Microtubule destabilizing agents are a critical class of compounds in cell biology research and drug development, functioning by inhibiting microtubule polymerization and promoting depolymerization. These agents target specific binding sites on tubulin, primarily the vinca domain and colchicine binding site, leading to disruption of mitotic spindle formation and cell cycle arrest at G2/M phase [62]. Within the context of microtubule lattice dynamics characterization research, understanding the specific mechanisms, efficacies, and limitations of these compounds is fundamental for experimental design and data interpretation. This technical support center provides essential resources for researchers investigating these powerful chemical tools.
Mechanisms of Action and Binding Sites
Microtubule destabilizing agents exert their effects through binding to distinct sites on the tubulin heterodimer. The table below summarizes the primary binding sites and molecular mechanisms of common destabilizing agents.
Table 1: Microtubule Destabilizing Agents: Binding Sites and Mechanisms
| Agent Class | Primary Binding Site | Molecular Mechanism | Key Structural Features |
|---|---|---|---|
| Vinca Alkaloids (Vincristine, Vinblastine) | Vinca Domain | Suppresses microtubule dynamics, promotes depolymerization, spiral tubulin structures [63] | Complex indole alkaloids |
| Colchicine-Site Binders (Combretastatin A-4, novel agents like "Hit22") | Colchicine Binding Site [62] | Inhibits tubulin polymerization, prevents microtubule assembly [62] | Three H-bond acceptors, one H-bond donor, two hydrophobic centers, one planar group [62] |
| Eribulin | Vinca Domain (distinct stoichiometry) [63] | Suppresses microtubule dynamics without extensive depolymerization [63] | Synthetic halichondrin analog |
The following diagram illustrates the cellular signaling pathways affected by microtubule destabilization, which lead to diverse functional outcomes.
Figure 1: Signaling Pathways in Microtubule Destabilization. This diagram outlines key cellular consequences of microtubule disruption, including mitotic arrest and cytoskeletal remodeling via the GEF-H1/RhoA pathway [63].
Troubleshooting Guides
FAQ: Addressing Common Experimental Challenges
Q1: My microtubule destabilizing agent is causing unexpected cytotoxicity in non-mitotic cells. What could be the reason? Microtubule destabilizers affect processes beyond mitosis, including intracellular transport, cell migration, and signal transduction [64]. Cytotoxicity in non-dividing cells may result from disruption of these essential functions. Consider:
- Dose Optimization: Test a range of concentrations (e.g., low nM to µM) to establish a therapeutic window.
- Exposure Time: Reduce treatment duration if possible.
- Agent Selection: Explore agents with purported cancer cell specificity, such as CMPD1, which was shown to preferentially induce mitotic defects in breast cancer cells over non-transformed normal cells at low concentrations (10 nM) [42].
Q2: I am observing inconsistent anti-proliferative effects with colchicine-site agents across different cell lines. How should I proceed? Variable responses are common due to factors like tubulin isotype expression and drug efflux pumps [62]. For systematic investigation:
- Confirm Target Engagement: Perform tubulin polymerization assays to verify the compound is effectively inhibiting microtubule assembly in your specific cell models [62].
- Quantitate Efficacy: Establish dose-response curves (IC₅₀ values) for anti-proliferative activity. For example, the colchicine-site agent "Hit22" exhibited an IC₅₀ of 3.93 µM in H1299 lung cancer cells [62].
- Check for Resistance Mechanisms: Analyze expression of class III β-tubulin and P-glycoprotein, which are associated with resistance to other microtubule-targeting agents like taxanes [42].
Q3: Why does a low dose of a vascular disrupting agent (e.g., Combretastatin A-4) appear to stabilize tumor vasculature in my model, contrary to its known disruptive function? The effects of colchicine-site binders can be dose-dependent. At low doses, agents like Combretastatin A-4 can induce pericyte phenotype switching, promoting a mature, contractile pericyte phenotype that stabilizes vasculature, improves perfusion, and reduces hypoxia [63]. At higher doses, the same agent causes rapid vascular disruption and collapse. Carefully re-evaluate your dosing regimen relative to the intended pharmacological outcome.
Q4: How can I enhance the efficacy of a microtubule destabilizer in a resistant cancer cell model? Combination strategies can overcome resistance. Recent research demonstrates that inhibiting the p38-MK2 signaling pathway sensitizes cancer cells to microtubule-targeting agents.
- Strategy: Co-administration of a specific MK2 inhibitor (e.g., MK2-IN-3) with a destabilizer like vinblastine [42].
- Experimental Protocol: Pre-treat cells with the MK2 inhibitor for 1-2 hours before adding the microtubule destabilizer. This approach allows for using a subclinical concentration of the destabilizer while still achieving significant mitotic arrest and cell death [42].
Quantitative Efficacy Profiling
The following table provides a comparative summary of quantitative data for selected microtubule destabilizing agents, crucial for experimental planning and benchmarking.
Table 2: Quantitative Efficacy Profile of Microtubule Destabilizing Agents
| Agent Name | Reported ICâ‚…â‚€ (Proliferation) | Cell Line / Model | Key Functional Outcomes |
|---|---|---|---|
| Hit22 (Novel Colchicine-site inhibitor) | 3.93 µM [62] | H1299 (Human Lung Cancer) [62] | Tubulin polymerization inhibition; G2/M cell cycle arrest; in vivo tumor growth suppression [62] |
| Vincristine (VCR) | Not explicitly quantified (Enhanced platelet yields) [65] | hiPSC-derived Megakaryocyte Lines (imMKCLs) [65] | Promotes proplatelet formation; reduces microtubule content in derived platelets [65] |
| CMPD1 (Dual p38-MK2/Microtubule inhibitor) | 10 nM (induced irreversible mitotic defects) [42] | MDA-MB-231 (Triple-Negative Breast Cancer) [42] | Rapid microtubule depolymerization; inhibits tumor growth, migration, and invasion [42] |
| Eribulin | Clinical response in <20% of pre-treated metastatic breast cancer patients [42] | Preclinical Breast Cancer Models / Clinical Trials [63] [42] | Tumor vascular remodeling; improved perfusion; enhanced immune cell infiltration (NK cells) [63] |
Standard Experimental Protocols
Protocol 1: In Vitro Tubulin Polymerization Assay
Purpose: To directly evaluate the microtubule destabilizing activity of a candidate compound.
- Reagent Preparation: Prepare purified tubulin protein (e.g., from bovine brain) in a polymerization buffer containing GTP [62].
- Baseline Measurement: Load tubulin solution into a pre-warmed cuvette in a spectrophotometer. Monitor turbidity at 350 nm to establish a baseline polymerization rate.
- Compound Addition: Add the test compound (dissolved in DMSO) to the tubulin solution. A final DMSO concentration of ≤1% is recommended. Include a vehicle control (DMSO only).
- Kinetic Analysis: Continuously monitor the absorbance at 350 nm over 60-90 minutes at 37°C. A successful destabilizing agent will show a significant reduction in the rate and extent of turbidity increase compared to the control, indicating inhibited polymerization [62].
Protocol 2: Analysis of Mitotic Arrest via Live-Cell Imaging
Purpose: To dynamically assess the compound's ability to induce mitotic arrest. This is a hallmark of anti-mitotic agents.
- Cell Preparation: Seed cancer cells (e.g., MDA-MB-231) expressing a fluorescent histone marker (e.g., H2B-GFP) into a multi-well imaging plate.
- Compound Treatment: Treat cells with the destabilizing agent at the desired concentration. Include a negative control (vehicle) and a positive control (e.g., 100 nM vinblastine).
- Image Acquisition: Place the plate in a live-cell imaging system maintained at 37°C and 5% CO₂. Acquire images every 5-10 minutes for 24-48 hours.
- Data Analysis: Track individual cells. A cell is considered arrested in prometaphase if it remains in a rounded state with condensed chromosomes for a significantly prolonged duration (e.g., >10 hours) compared to control cells, which typically complete mitosis within 30-60 minutes [42].
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Microtubule Dynamics Studies
| Reagent / Material | Function / Application | Specific Examples / Notes |
|---|---|---|
| EB1-GFP | Live-cell marker for tracking growing microtubule plus ends; essential for quantifying microtubule dynamics [66]. | Used in lattice light-sheet microscopy (LLSM) to map microtubule growth trajectories in 3D with high precision [66]. |
| Anti-Polyglutamylation Antibody | Detects tubulin post-translational modification (polyE chains) that regulates microtubule severing and stability [67]. | Critical for studies linking microtubule dynamics to neuronal remodeling; used in immunohistochemistry and Western blotting [67]. |
| imMKCLs (expandable hiPSC-derived megakaryocytic cell lines) | Model system for studying microtubule role in platelet biogenesis and screening for compounds affecting this process [65]. | Used to identify VCR as a promoter of proplatelet formation, linking microtubule destabilization to a specialized cellular differentiation outcome [65]. |
| p38-MK2 Pathway Inhibitors (e.g., CMPD1, MK2-IN-3) | Tool compounds to investigate synergy with microtubule destabilizers and to overcome potential resistance [42]. | CMPD1 is a dual-target inhibitor; MK2-IN-3 is a more specific inhibitor for combination studies [42]. |
In Vitro Tubulin Polymerization vs. Cellular Microtubule Content Assays
Within research characterizing microtubule lattice dynamics, a fundamental challenge is selecting and correctly implementing the appropriate assay to interrogate the effects of chemical compounds or cellular proteins on tubulin. Researchers must choose between reductionist, cell-free in vitro tubulin polymerization assays and more complex cellular microtubule content assays. The former offers mechanistic clarity by studying purified components, while the latter provides essential biological context. This technical support center is designed to help you troubleshoot specific issues, understand key methodological differences, and select the right tools for your experimental goals in drug discovery and basic research.
FAQs: Understanding the Core Assays
What is the fundamental difference between these two assay types?
The core difference lies in the experimental system and the type of information they provide.
- In Vitro Tubulin Polymerization Assay: This is a biochemical assay conducted in a cell-free environment using purified tubulin protein. It directly measures the ability of a test compound to promote or inhibit the assembly of tubulin into microtubules under defined buffer conditions. The readout is a kinetic curve of the polymerization reaction, allowing for the calculation of parameters like lag time, polymerization rate, and final polymer mass [68] [69] [70].
- Cellular Microtubule Content Assay: This is a cell-based assay that measures the amount and organization of polymerized tubulin within intact cells. It indirectly assesses a compound's effect on the microtubule cytoskeleton by quantifying staining intensity and morphological changes after fixation and immunostaining. It reveals how a compound influences microtubules in a biologically complex environment, including effects mediated through cellular pathways rather than direct binding [70].
When should I choose an in vitro polymerization assay?
An in vitro assay is the most appropriate choice for the following objectives:
- Confirming Direct Target Engagement: To unequivocally determine if your compound binds directly to tubulin and alters its polymerization kinetics [69].
- Mechanistic Studies: To study the biophysical properties of tubulin polymerization itself, or to characterize the mechanism of action of a known tubulin-binding compound (e.g., stabilizer vs. destabilizer) [68].
- Initial High-Throughput Screening: For the cost-effective screening of large compound libraries due to its simpler setup and homogenous format [70].
- Determining Potency (ECâ‚…â‚€/ICâ‚…â‚€): To generate quantitative data on a compound's efficacy in promoting or inhibiting polymerization from purified components [69].
When is a cellular microtubule assay necessary?
A cellular assay is indispensable in these scenarios:
- Evaluating Cell Permeability: To determine if a compound active in the in vitro assay can cross the cell membrane to reach its intracellular target [70].
- Assessing Functional Consequences: To link tubulin modulation to phenotypic outcomes like cell cycle arrest (G2/M phase), mitotic spindle defects, or changes in cell morphology [70].
- Differentiating Mechanism in a Cellular Context: To distinguish between tubulin stabilizers and destabilizers based on their direct effect on the cellular microtubule network [70].
- Profiling Off-Target Effects: A cellular active compound that fails in the in vitro assay may have an indirect mechanism of action, working through a different cellular target that secondarily affects microtubules.
My compound is active in vitro but inactive in cells. What are the potential causes?
This common discrepancy can typically be traced to a few key issues:
- Poor Cell Permeability: The compound may be unable to efficiently cross the cell membrane.
- Efflux by Transporters: It may be actively pumped out of cells by efflux transporters like P-glycoprotein [70].
- Intracellular Metabolism: The compound could be metabolized and inactivated inside the cell before it reaches its target.
- Binding to Serum Proteins: In cell culture, serum proteins in the medium can bind the compound, reducing its free, active concentration.
My compound is active in cells but inactive in vitro. What does this mean?
This result strongly suggests that the compound does not act by directly binding to tubulin. Its effect on cellular microtubules is likely indirect, potentially through:
- Regulation of Microtubule-Associated Proteins (MAPs): It may activate or inhibit proteins that stabilize or destabilize microtubules.
- Altering Signaling Pathways: It could affect kinase/phosphatase pathways that regulate microtubule dynamics.
- Transcriptional or Translational Control: It might downregulate tubulin expression or upregulate a microtubule-destabilizing factor.
Troubleshooting Guides
Issue 1: Poor or No Polymerization in the In Vitro Assay
| Possible Cause | Diagnostic Experiments | Recommended Solution |
|---|---|---|
| Incorrect Tubulin Quality/Handling | Check tubulin purity via SDS-PAGE. Ensure it was kept on ice and spun briefly before use to remove aggregates [71]. | Use fresh, high-purity tubulin from a reliable supplier. Avoid multiple freeze-thaw cycles. Perform a quick spin before use [71]. |
| Suboptimal Buffer Conditions | Verify pH of PIPES buffer (typically 6.9) and concentration of critical components like MgClâ‚‚ and GTP [72] [71]. | Prepare fresh assay buffer. Use a glutamate-based buffer as an alternative, which can promote polymerization and be "tuned" by concentration [71]. |
| Inadequate Polymerization Enhancer | Run a positive control (e.g., Paclitaxel) with and without glycerol. | Include a polymerization enhancer like glycerol (e.g., 10-15%) in the reaction mix [72] [70]. |
| Temperature Fluctuation | Monitor plate temperature in the reader to ensure a stable 35-37°C. | Pre-warm the plate reader and all buffer components. Use a thermal plate reader with a heated lid to prevent condensation [68]. |
Issue 2: High Background Signal in Cellular Tubulin Staining
| Possible Cause | Diagnostic Experiments | Recommended Solution |
|---|---|---|
| Insufficient Washing or Blocking | Compare staining with increased wash steps and different blocking buffer concentrations. | Include rigorous washing after each antibody incubation step. Use a commercial blocking buffer and optimize incubation time (e.g., 1 hour) [70]. |
| Antibody Concentration Too High | Perform an antibody titration experiment to find the optimal dilution. | Titrate both primary and secondary antibodies. Using a highly cross-adsorbed secondary antibody can reduce non-specific binding. |
| Fixation or Permeabilization Artifacts | Test different fixatives (e.g., formaldehyde vs. methanol) and permeabilization agents (e.g., Triton X-100 vs. saponin). | Standardize on 4% formaldehyde for fixation. Use a consistent, optimized permeabilization step (e.g., 20 minutes with a commercial buffer) [70]. |
| Autofluorescence | Image untreated, unstained cells under the same channels. | Include controls without antibodies. Choose fluorescent dyes with emission spectra outside the autofluorescence range. Use a dye-quenching solution if necessary. |
Issue 3: Inconsistency Between Technical Replicates in the In Vitro Assay
| Possible Cause | Diagnostic Experiments | Recommended Solution |
|---|---|---|
| Inconsistent Tubulin Thawing/Aliquoting | Compare polymerization curves from different tubulin aliquots. | Always prepare a fresh, master mix of tubulin for all reactions in an experiment. Avoid repeated freeze-thaw cycles [68] [71]. |
| Improper Plate Selection | Check the plate manufacturer's recommendation for low-binding surfaces. | Use 96-well or 384-well plates with a non-binding surface to prevent tubulin from sticking to the well walls [72] [70]. |
| Edge Effects in Plate Reader | Compare polymerization kinetics in edge wells versus center wells. | Use a thermostatted plate reader with a controlled chamber. Pre-warm the plate and consider using only the inner wells of the plate. |
Table 1: Key Parameters for In Vitro Tubulin Polymerization Assays. Data is compiled from standard protocols using mammalian brain tubulin [72] [68] [70].
| Parameter | Typical Range | Description & Impact |
|---|---|---|
| Tubulin Concentration | 2-3 mg/mL | A critical determinant of polymerization kinetics and final polymer mass. |
| GTP Concentration | 1 mM | Essential cofactor for polymerization; its hydrolysis drives dynamics. |
| Incubation Temperature | 35-37°C | Polymerization is highly temperature-dependent and must be strictly maintained. |
| Positive Control (Stabilizer) | 3-10 μM Paclitaxel | Serves as a benchmark for robust polymerization [72]. |
| Positive Control (Destabilizer) | 3-10 μM Colchicine/Nocodazole | Serves as a benchmark for inhibited polymerization [72] [70]. |
| Polymerization Kinetics Measurement | 60 minutes | Standard duration to capture the lag phase, growth phase, and plateau. |
Table 2: Comparison of Tubulin Polymerization Assay Methodologies.
| Feature | In Vitro (Turbidity) | In Vitro (Fluorescence) | Cellular (High-Content) |
|---|---|---|---|
| Principle | Light scattering by polymerized structures [71] | Enhanced fluorescence upon dye binding to polymer (e.g., DAPI) [70] | Immunofluorescence intensity & morphology [70] |
| Key Reagents | Purified tubulin, GTP, buffer | Purified tubulin, GTP, fluorescent dye (e.g., DAPI), buffer | Cultured cells, fixation/permeabilization reagents, tubulin antibodies [70] |
| Throughput | High | High | Medium (requires image analysis) |
| Direct vs. Indirect | Direct measure of polymerization | Direct measure of polymer mass | Indirect measure of polymer content |
| Information Gained | Kinetic parameters (lag, rate, mass) | Kinetic parameters (lag, rate, mass) | Network organization, phenotypic effects |
| Cost | Low | Low | Medium to High |
Experimental Protocols
Detailed Protocol: In Vitro Tubulin Polymerization (Fluorescence-Based)
This protocol uses a fluorescence-based method, which is more sensitive than turbidity and is commonly available in most labs with a plate reader [72] [70].
Workflow: In Vitro Tubulin Polymerization Assay
Materials & Reagents:
- Purified tubulin (e.g., from porcine brain, >97% pure) [72] [68].
- Tubulin Polymerization Buffer: 80 mM PIPES pH 6.9, 2 mM MgClâ‚‚, 0.5 mM EGTA, 1 mM GTP [72] [70].
- Fluorescent Reporter: 10 μM (e.g., DAPI at 6.3 μM) [72] [70].
- Polymerization Enhancer: 15% glycerol [72].
- Test compounds and controls (e.g., 3 μM Paclitaxel and Colchicine) [72].
- Non-binding 96-well black wall plate [70].
- Fluorescence plate reader capable of maintaining 37°C (e.g., Varioskan Flash, Spectramax Gemini) [72] [70].
Step-by-Step Procedure:
- Preparation: Thaw all components on ice. Prepare a tubulin master mix on ice by reconstituting lyophilized tubulin or diluting a stock in the provided polymerization buffer to a final concentration of 2-3 mg/mL. Include the fluorescent reporter (DAPI) and glycerol in the master mix. Briefly centrifuge the tubulin mix at high speed (e.g., 20,000 x g for 5 minutes at 4°C) to remove any aggregates, and use the top 90% of the supernatant [71] [70].
- Plate Setup: Dispense the test compounds and controls into a pre-warmed 96-well plate. The final volume of compounds should be small compared to the tubulin mix volume (e.g., 1-2% v/v) to avoid dilution effects. A typical control setup includes:
- Negative Control: Buffer with DMSO vehicle (e.g., 0.1% DMSO).
- Positive Control (Stabilizer): 3 μM Paclitaxel.
- Positive Control (Destabilizer): 3 μM Colchicine [72].
- Initiate Polymerization: Add the pre-warmed tubulin master mix to each well. Mix thoroughly by pipetting up and down several times, taking care not to introduce bubbles.
- Kinetic Measurement: Immediately place the plate into the pre-warmed (37°C) plate reader. Program the reader to take fluorescence readings (e.g., Ex/Em 360/450 nm for DAPI) every minute for 60-90 minutes [72] [70].
- Data Analysis: Plot the fluorescence intensity versus time for each well. The data is often presented as the Area Under the Curve (AUC) for the entire kinetic run or by analyzing the lag time, polymerization rate (slope of the linear phase), and final extent of polymerization [72] [70].
Detailed Protocol: Cellular Microtubule Content (High-Content Analysis)
This protocol outlines a quantitative high-content analysis method to distinguish tubulin stabilizers from destabilizers in a 384-well format [70].
Workflow: Cellular Microtubule Content Assay
Materials & Reagents:
- Cell line (e.g., A549 or HCT116 cells) [70].
- 384-well clear bottom black-wall poly-D-lysine coated microplates [70].
- Fixation Solution: 4% formaldehyde in DPBS (with Ca²⺠and Mg²âº) [70].
- Permeabilization Buffer (commercial, e.g., from ThermoFisher) [70].
- Blocking Buffer (commercial, e.g., from ThermoFisher) [70].
- Primary Antibody: Anti-α-Tubulin (e.g., CBL270 from Millipore) [70].
- Secondary Antibody: Anti-rat conjugated to Alexa488 [70].
- Nuclear Stain: Hoechst 33342 [70].
- High-content imaging system (e.g., GE InCell 2000) [70].
Step-by-Step Procedure:
- Cell Plating: Harvest and seed cells into 384-well plates at a low density (e.g., 2,000 cells per well) and incubate overnight at 37°C in a CO₂ incubator [70].
- Compound Treatment: Treat cells with a range of concentrations of the test compounds for a defined period (e.g., 3, 6, or 18 hours at 37°C). Include controls like DMSO (vehicle), Paclitaxel (stabilizer), and Nocodazole (destabilizer) [70].
- Fixation and Permeabilization: After treatment, carefully aspirate the medium and fix the cells by adding 4% formaldehyde for 30 minutes at room temperature. Wash twice with DPBS, then permeabilize the cells with Permeabilization Buffer for 20 minutes, followed by two more washes [70].
- Immunostaining: Incubate cells with Blocking Buffer for 1 hour. Without washing, add the primary antibody (anti-α-Tubulin, diluted 1:125 in blocking buffer) and incubate overnight at 4°C. Wash twice with DPBS, then add the secondary antibody (anti-rat Alexa488, diluted 1:500) along with Hoechst 33342 (4 μg/mL) and incubate for 3 hours at room temperature in the dark. Perform final two washes with DPBS [70].
- Image Acquisition and Analysis: Acquire fluorescent images on a high-content platform using a 20x objective. Use the analysis software to segment the nuclei and cytoplasm. The key measurement is the total tubulin staining intensity in the cytoplasmic region. Tubulin stabilizers will increase this intensity, while destabilizers will decrease it. This allows for the generation of concentration-response curves and the distinction between the two mechanisms [70].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Microtubule Assays.
| Item | Function | Example & Notes |
|---|---|---|
| Purified Tubulin | The core protein component for in vitro assays. | Porcine brain tubulin is common and cost-effective [68] [69]. Recombinant or cancer cell line-derived tubulin is also available for specific applications [69]. |
| Tubulin Polymerization Kits | Provide a complete, optimized system for in vitro assays. | Available from specialty suppliers (e.g., Cytoskeleton, Inc.), these kits include tubulin, optimized buffer, GTP, fluorescent reporter, and controls [72] [69] [70]. |
| Microtubule-Stabilizing Agents | Positive controls for polymerization/stabilization. | Paclitaxel (Taxol) is the canonical stabilizer. Discodermolide and newer-generation taxoids can also be used [68]. |
| Microtubule-Destabilizing Agents | Positive controls for depolymerization/destabilization. | Colchicine (binds soluble tubulin), Nocodazole (binds tubulin dimer, reversible), and Vinblastine (induces paracrystals) [70]. |
| Tubulin Antibodies | Critical for detecting and visualizing microtubules in cells. | Anti-α-Tubulin and Anti-β-Tubulin monoclonal antibodies are widely used for immunofluorescence [70]. |
| Fluorescent Dyes & Secondaries | For detection in cellular and fluorescence-based in vitro assays. | DAPI binds polymerized tubulin in vitro [70]. Alexa Fluor-conjugated secondary antibodies provide high signal for cellular imaging [70]. |
| Live-Cell Imaging Reagents | For dynamic studies of microtubules in living cells. | CellLight Tubulin-GFP reagents (baculovirus-based) for labeling microtubules in live cells [73]. |
Lattice Dynamics as Early Biomarkers in Neurodegenerative Disease
This technical support resource is designed for researchers characterizing microtubule lattice dynamics in the context of neurodegenerative disease. The center provides targeted troubleshooting guides, detailed experimental protocols, and reagent information to address common challenges in this emerging field. The content is framed within a research thesis that hypothesizes specific, quantifiable alterations in microtubule lattice spacing precede traditional histopathological markers in neurodegeneration, positioning lattice dynamics as a novel class of early biomarker.
Frequently Asked Questions (FAQs) & Troubleshooting
Q1: Our in vitro assays show inconsistent microtubule lattice behavior. What are the primary factors affecting lattice stability?
A1: Inconsistent lattice stability often stems from competing molecular forces. Your system may contain both microtubule "expanders" and "compactors" whose effects are concentration-dependent.
- Root Cause: Many buffers and experimental conditions contain agents that implicitly compact or expand the lattice. The competition between these forces defines the final lattice spacing state [3].
- Solution:
- Audit your reagents: Identify potential expanders (e.g., paclitaxel) and compactors (e.g., Doublecortin/DCX) in your protocol.
- Systematic Titration: Perform a matrix of experiments where you titrate the suspected expander against the compactor. The relative concentration, not the absolute concentration of either, often determines the outcome [3].
- Control the Nucleotide State: Remember that GTP-like lattices are often expanded, while GDP-lattices are typically compacted. Use non-hydrolyzable GTP analogs (e.g., GMPCPP) to control for this variable [3].
Q2: When using cryo-EM to measure lattice spacing, what is the expected difference between expanded and compacted states, and how can I ensure I'm detecting a biologically relevant change?
A2: The difference is subtle but statistically significant and has functional consequences.
- Expected Values: Measurements cluster into two primary states [3]:
- Expanded State: 83.5 ± 0.2 Å
- Compacted State: 81.7 ± 0.1 Å
- This represents a difference of 2.3 ± 0.2 %.
- Troubleshooting: Ensure your image processing and analysis pipelines are calibrated to detect sub-nanometer changes. Use internal standards and validate findings against a known expanded (e.g., GMPCPP) or compacted (e.g., DCX) control in your system.
Q3: How can I bridge the gap between molecular-level lattice measurements and tissue-level pathology in my disease models?
A3: This is a central multiscale challenge. We recommend employing quantitative spatial statistics on tissue imaging data to infer underlying aggregation mechanisms.
- Recommended Workflow:
- Image Analysis: From immunohistochemistry or immunofluorescence images of patient or model tissue, derive spatial statistics. Key metrics include [74]:
- Nearest Neighbour Distance (NND) Distribution: The probability distribution of distances from an aggregated cell to its nearest affected neighbor.
- Radial Distribution Function (RDF): The average fraction of pathologically affected cells at a distance
rfrom another affected cell.
- Model Comparison: Compare your experimental spatial patterns to the fingerprints of different mechanistic models. For instance [74]:
- A random, uncorrelated spatial pattern of pathology suggests a cell-autonomous process (e.g., spontaneous nucleation within independent cells).
- Clustered, spatially correlated patterns suggest a propagation-dominated process (e.g., cell-to-cell spread).
- Image Analysis: From immunohistochemistry or immunofluorescence images of patient or model tissue, derive spatial statistics. Key metrics include [74]:
Q4: What are the most promising translational approaches for measuring lattice dynamics in vivo for biomarker validation?
A4: While direct lattice measurement in vivo remains a challenge, proxy methods are in development.
- Primary Approach: MT-Targeted PET Imaging. The field is developing radiotracers that selectively bind to destabilized microtubules. The tracer
[11C]MPC-6827demonstrates high specificity for destabilized MTs and excellent brain uptake, allowing for non-invasive, in vivo visualization of MT integrity [44]. - Correlative Biomarkers: Monitor post-translational modifications (PTMs) of tubulin, such as acetylation and detyrosination. The writers of these PTMs, like the acetyltransferase αTAT1, are regulated by lattice spacing, making the PTMs themselves indirect reporters of lattice state [3].
Table 1: Key Quantitative Metrics in Microtubule Lattice Dynamics Research
| Metric | Typical Value / Range | Significance / Interpretation | Experimental Context |
|---|---|---|---|
| Lattice Spacing (Expanded) | 83.5 ± 0.2 Å | GTP-like state; associated with kinesin-1 binding, paclitaxel [3] | Cryo-EM measurement [3] |
| Lattice Spacing (Compacted) | 81.7 ± 0.1 Å | GDP-state; associated with DCX, tau, EB proteins [3] | Cryo-EM measurement [3] |
| Lattice Spacing Difference | 2.3 ± 0.2 % | Represents the "accordion-like" transition that reorganizes tubulin contact surfaces [3] | Calculated from cryo-EM data [3] |
| Spatial Agreement Measure (SAM) | >77% accuracy | Quantifies how well a model-predicted spatial cell distribution matches an observed biopsy [75] | Used to validate agent-based models of tumor-immune interactions [75] |
Detailed Experimental Protocols
Protocol 1: In Vitro Microtubule Lattice Buckling Assay
Purpose: To directly observe the effects of molecular agents (e.g., expanders vs. compactors) on the physical properties of pre-formed microtubules using light microscopy [3].
Workflow Diagram: In Vitro Buckling Assay
Reagents & Materials:
- Purified tubulin
- Non-hydrolyzable GTP analog (GMPCPP) for stable caps
- Flow chamber for immobilization
- Test compounds (e.g., paclitaxel, docetaxel, DCX)
- Light microscope with high-resolution optics
Step-by-Step Method:
- Polymerize Double-Capped Microtubules: Assemble microtubules with stable, short caps of GMPCPP-tubulin at both ends, leaving a long central segment of GDP-lattice.
- Immobilize: Anchor these microtubules to the cover glass of a flow chamber.
- Introduce Compound: Flow in the test compound (e.g., paclitaxel, DCX, or both at varying ratios) and incubate.
- Induce Buckling: Apply axial compressive force to the microtubules. This can occur naturally from thermal forces or be induced.
- Image and Quantify: Observe and record the buckling dynamics. The buckling wavelength is a direct physical readout of the microtubule's mechanical stiffness, which is influenced by its lattice spacing. An expanded lattice will have a different buckling wavelength than a compacted one [3].
Protocol 2: Spatial Analysis of Protein Aggregation in Tissue
Purpose: To determine whether a pathological protein aggregation pattern in tissue is driven by cell-autonomous or propagation mechanisms, using spatial statistics [74].
Workflow Diagram: Spatial Pattern Analysis
Reagents & Materials:
- Tissue sections (e.g., from animal models or post-mortem brain)
- Antibodies for specific pathological proteins (e.g., phosphorylated tau, α-synuclein)
- Whole-slide scanner or high-content microscope
- Image analysis software (e.g., ImageJ, QuPath, custom scripts in Python/R)
Step-by-Step Method:
- Image Acquisition: Generate high-resolution digital images of immunostained tissue sections.
- Cell Identification and Segmentation: Use software to identify and mark the position of every cell (or a representative sample) and classify its state as "healthy" or "aggregated" based on staining intensity and morphology.
- Calculate Spatial Statistics:
- Nearest Neighbour Distance (NND): For every aggregated cell, compute the distance to its nearest aggregated neighbor. Plot the distribution of these distances.
- Radial Distribution Function (RDF): For every aggregated cell, calculate the density of other aggregated cells at increasing distances
rfrom it. Average this across all cells to getg_norm(r).
- Interpretation:
- A NND distribution that fits a Poisson point process and an RDF that is flat and equal to the overall fraction of affected cells indicates a random, cell-autonomous process [74].
- A NND distribution shifted towards shorter distances and an RDF that shows a peak at short distances indicates spatial clustering consistent with propagation [74].
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for Microtubule Lattice Dynamics Research
| Reagent / Tool | Category | Key Function in Research | Example & Notes |
|---|---|---|---|
| Paclitaxel (Taxol) | Microtubule Stabilizer / Expander | Nucleates expanded lattices; used to experimentally induce lattice expansion and study competition with compactors [3]. | European Pharmacopoeia (Cat#Y0000698) [3]. Effects are concentration-dependent and can be overridden by high levels of compactors. |
| Doublecortin (DCX) | Neuronal MAP / Compactor | Nucleates compacted lattices; a key compactor used to oppose the effects of expanders like paclitaxel [3]. | Recombinant protein. A critical tool for manipulating lattice spacing toward the compacted state. |
| GMPCPP | Non-hydrolyzable GTP Analog | Creates stable, expanded microtubule caps; essential for controlling nucleotide state in polymerization and buckling assays [3]. | A GTP analog that mimics the GTP-state, leading to expanded lattices that are resistant to depolymerization. |
| [11C]MPC-6827 | PET Radiotracer | Selectively binds destabilized microtubules; enables non-invasive, in vivo imaging of microtubule integrity in the brain [44]. | Emerging translational tool for connecting basic lattice dynamics to whole-brain pathology in living organisms. |
| TREM2-Activating Antibodies | Therapeutic / Probe | Modulates microglial function; used to investigate the role of microglia in clearing aggregates in a tauopathy context [76]. | e.g., AL002 (Alector). While targeting microglia, it is relevant for studying non-cell-autonomous effects on neuronal health. |
Conceptual Framework & Signaling Pathways
Diagram: The Interplay of Lattice Spacing, Physiology, and Pathology
Frequently Asked Questions (FAQs)
Q1: What specific biological target does the [11C]MPC-6827 radiotracer bind to, and what does its uptake indicate? A1: [11C]MPC-6827 is a high-affinity, selective binder of the β-tubulin subunit within microtubules [77] [78]. Its uptake has an inverse correlation with microtubule stability. Higher radioactive signal indicates a greater presence of destabilized microtubules, while lower uptake is observed in regions with stable microtubules [44] [77].
Q2: How does the presence of tau protein affect [11C]MPC-6827 binding? A2: Research in tau knockout (KO) mouse models shows that the absence of tau protein leads to significantly higher [11C]MPC-6827 uptake compared to wild-type mice [77]. This suggests that radiotracer binding does not require the presence of aggregated tau and may, in fact, better reflect the absence of functional tau that normally stabilizes microtubules. This makes it particularly useful for probing early-stage dysfunction before significant tau pathology develops [77].
Q3: What are the key binding and metabolic properties that make [11C]MPC-6827 suitable for CNS PET imaging? A3: [11C]MPC-6827 exhibits ideal characteristics for a central nervous system (CNS) radiopharmaceutical, as shown in the table below [78]:
Table 1: Key Binding and Metabolic Properties of [11C]MPC-6827
| Property | Reported Value | Experimental Context |
|---|---|---|
| Lipophilicity (LogP) | 2.9 | In vitro measurement in PBS buffer/1-octanol |
| Dissociation Constant (Kd) | 15.59 nM | In vitro autoradiography on mouse brain tissue |
| Maximum Binding Capacity (Bmax) | 11.86 fmol/mg | In vitro autoradiography on mouse brain tissue |
| Serum Stability | >95% (at 3 hours) | Ex vivo incubation in human serum |
| Brain Metabolic Stability | >95% | In vivo study in rat brain samples |
| Specificity (Blocking) | >70% decrease | In vivo pretreatment with non-radioactive MPC-6827 |
Q4: What is the established radiochemistry protocol for producing [11C]MPC-6827? A4: The synthesis is performed using a automated radiochemistry module (e.g., GE-FXC) [77] [78]:
- Precursor: Desmethyl-MPC-6827 (0.8-1 mg).
- Methylation: Reaction with [11C]Methyl Iodide ([11C]MeI) in a solution of NaOH/DMF at 80°C for 3-5 minutes.
- Purification: Semi-preparative HPLC using a C18 column with a mobile phase of acetonitrile and 0.1 M aqueous ammonium formate (pH 6.0-6.5).
- Formulation: The purified product is trapped on a C18 SepPak cartridge and eluted with ethanol into sterile saline, resulting in a final formulation of 10% ethanol in saline. The entire process, from [11C]CO2 to final sterile product, is completed within approximately 50 minutes [77] [78].
Troubleshooting Guides
Issue: Unexpectedly Low Radiotracer Uptake in Brain Tissue
Table 2: Troubleshooting Low Radiotracer Uptake
| Possible Cause | Suggested Experiments | Expected Outcome |
|---|---|---|
| Microtubule Over-stabilization | Treat a subject group with a MT-stabilizing agent (e.g., EpoD). Perform pre-treatment blocking studies with non-radioactive MPC-6827 [77]. | Uptake should decrease significantly with stabilizer and blocking agent, confirming on-target binding. |
| Poor Radiochemical Purity | Perform QC-HPLC analysis immediately after synthesis to confirm radiochemical purity and specific activity [78]. | Resolving synthesis issues should restore expected uptake levels. |
| Loss of Tubulin Binding Sites | Conduct post-mortem analysis of brain tissue using Western blot or capillary electrophoresis to quantify β-tubulin levels [77]. | A correlation between low Bmax values from binding assays and low β-tubulin protein levels confirms target loss. |
Issue: High Non-Specific Binding in Pre-Clinical Models
- Validate with In Vitro Cell Uptake Assay:
- Culture: Use SH-SY5Y neuronal cells (0.5 × 10^6 cells/well).
- Pre-treatment: Incubate cells for 3 hours with:
- Stabilizing agents (1.0 µM Paclitaxel or EpoD).
- Destabilizing agents (1.0 µM Vinblastine or Mertasine).
- Blocking agent (1.0 µM non-radioactive MPC-6827).
- Tracer Incubation: Add [11C]MPC-6827 (0.074 MBq/well) and incubate for 30 minutes at room temperature.
- Measurement: Measure radioactive uptake in the cells.
- Expected Result: Uptake should be lower in stabilized cells and higher in destabilized cells compared to untreated controls. The blocking group should show significantly reduced uptake, confirming specificity [77].
Experimental Protocols
Protocol for Validating Target Engagement with MicroPET/CT
This protocol is used to demonstrate that in vivo radiotracer binding is specific to its microtubule target.
- Animal Preparation: Anesthetize and physiologically monitor the animal (e.g., wild-type mouse).
- Pre-treatment (Experimental Group): Administer a brain-penetrant microtubule-stabilizing drug (e.g., EpoD).
- Radiotracer Injection: Intravenously inject [11C]MPC-6827 via a tail or femoral vein catheter.
- Image Acquisition: Position the animal in the microPET/CT scanner. Acquire dynamic PET data for approximately 60-90 minutes, simultaneously with a low-dose CT scan for anatomical co-registration and attenuation correction.
- Image Analysis: Reconstruct the PET data and co-register with the CT images. Quantify the standardized uptake value (SUV) or binding potential (BP) in relevant brain regions.
- Validation: Compare the results with a control group that did not receive the stabilizing drug. A statistically significant decrease in uptake in the pre-treated group confirms specific binding to stabilized microtubules [77].
Protocol for Ex Vivo Biodistribution and Autoradiography
This protocol provides high-resolution, quantitative data on radiotracer distribution and binding post-imaging.
- Tissue Preparation: Following the in vivo PET scan, euthanize the animal and perfuse transcardially with ice-cold saline. Extract the brain and rapidly freeze it in optimal cutting temperature (OCT) compound.
- Sectioning: Cryosection the brain sagitally at a thickness of 20 µm and mount the sections on glass slides.
- In Vitro Incubation: Rehydrate the sections in PBS (pH 7.4) for 5 minutes. Incubate with a known concentration of [11C]MPC-6827 for 30 minutes at room temperature. For saturation binding experiments, use a range of concentrations.
- Exposure: Expose the dried sections to a phosphor imaging plate or film along with radioactive standards for several hours.
- Data Quantification: Scan the imaging plate and use the standards to convert optical density to radioactivity concentration (nCi/mg). Analyze regional binding and calculate binding parameters (Kd, Bmax) using appropriate software (e.g., ImageJ, PMOD) [78].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Microtubule PET Imaging
| Item | Function/Application | Example/Catalog |
|---|---|---|
| Tubulin Modulators (Stabilizers) | Pharmacological agents used to validate tracer specificity in vitro and in vivo. | Epothilone D (EpoD), Paclitaxel [77] |
| Tubulin Modulators (Destabilizers) | Pharmacological agents used to induce a high-uptake state for tracer validation. | Vinblastine, Mertasine [77] |
| Blocking Agent | Non-radioactive compound to confirm specific binding in competition assays. | Non-radioactive MPC-6827 hydrochloride [77] [78] |
| Tubulin Antibodies | For post-mortem validation of tubulin levels and post-translational modifications via Western blot. | β-Tubulin (Cat. # 2128), α-Tubulin (Cat. # 3873), Acetyl-α-Tubulin (Cat. # 5335T) [77] |
| Microtubule/Tubulin Assay Kit | For quantitative analysis of tubulin and microtubule content in tissue homogenates. | Cytoskeleton, Inc. MT/Tubulin Assay Kit (BK038) [77] |
| Radiochemistry Precursor | Essential starting material for the synthesis of [11C]MPC-6827. | Desmethyl MPC-6827 [77] [78] |
Signaling Pathways and Experimental Workflows
[11C]MPC-6827 Binding Mechanism
Diagram Title: Radiotracer Binding to Destabilized Microtubules
Experimental Workflow for Radiotracer Validation
Diagram Title: Workflow for Validating Microtubule PET Tracers
Cross-Species Validation of Microtubule-Targeting Therapies
Foundational Concepts: Microtubule Dynamics and Therapeutic Targeting
What are the core structural and dynamic properties of microtubules that make them effective therapeutic targets?
Microtubules are essential components of the cytoskeleton in eukaryotic cells, composed of αβ-tubulin heterodymers organized into protofilaments that form hollow cylindrical structures [79]. They display dynamic instability, characterized by stochastic growth and shrinkage at their ends, and lattice dynamics, where tubulin exchange occurs along the microtubule shaft [80]. These dynamic properties are crucial for cellular functions including cell division, intracellular transport, and maintenance of cell shape. Microtubule-Targeting Agents (MTAs) interfere with these dynamics, leading to cell cycle arrest and apoptosis, making them valuable tools in cancer therapy and other diseases [81] [79].
What are the primary classifications of Microtubule-Targeting Agents (MTAs) and their mechanisms of action?
MTAs are broadly classified into three categories based on their effect on microtubule polymerization, as detailed in the table below.
Table 1: Classification and Mechanisms of Microtubule-Targeting Agents
| Class | Mechanism of Action | Representative Agents | Primary Therapeutic Use |
|---|---|---|---|
| Microtubule Stabilizing Agents | Promote tubulin assembly and inhibit depolymerization; suppress dynamic instability [79]. | Taxanes (Paclitaxel, Docetaxel), Epothilones (Ixabepilone) [81] [39] | Various cancers (e.g., breast, prostate) [39] |
| Microtubule Destabilizing Agents | Inhibit tubulin polymerization, leading to microtubule disassembly [79]. | Vinca alkaloids (Vinblastine, Vincristine), Colchicine-site binders (Combretastatin A-4) [81] [39] | Various cancers (e.g., hematologic, thyroid) [39] |
| Microtubule Degradation Agents | Trigger tubulin degradation via the ubiquitin-proteasome system; a novel, pre-clinical class [39]. | — (No approved drugs yet) [39] | — (Under investigation for cancer) |
Experimental Protocols: Characterizing Lattice Dynamics and Drug Effects
A deep understanding of microtubule lattice behavior is fundamental for evaluating therapy efficacy. The following protocol and visualization outline a key method for analyzing intrinsic microtubule structure.
Segmented Subtomogram Averaging (SSTA) for Microtubule Lattice Analysis
This protocol is used to characterize the structural heterogeneity of individual microtubules, such as changes in the number and location of lattice "seams" (heterotypic interfaces) [82] [83].
Key Reagents:
- Microtubules: Assembled in vitro from purified tubulin or in cytoplasmic extracts (e.g., from Xenopus eggs) [82] [83].
- Decoration Protein: Kinesin motor-domains, which bind to every αβ-tubulin heterodimer to reveal the underlying lattice organization [82] [83].
- Software: IMOD and PEET for tomogram modeling and subtomogram averaging [82] [83].
Workflow Summary:
- Sample Preparation & Imaging: Decorate microtubules with kinesin motor-domains and acquire cryo-electron tomograms [82] [83].
- Modeling: Use
3dmod(from IMOD) to model the paths of individual protofilaments and the microtubule center throughout the tomogram [82] [83]. - Subvolume Extraction: Extract sub-volumes at each kinesin binding position to generate a full-length subtomogram average of the microtubule [82] [83].
- Segmentation and Analysis: Divide the full model into shorter segments. Calculate subtomogram averages for each segment independently to reveal local variations in lattice structure, such as seam location and the presence of holes [82] [83].
SSTA Workflow for Lattice Analysis
Protocol for Investigating Tau-Mediated Lattice Remodeling
Recent research reveals that the microtubule-associated protein Tau actively remodels the microtubule lattice, accelerating tubulin exchange [4]. The following assay investigates this phenomenon.
Key Reagents:
- Stabilized Microtubule Seeds: GMPCPP-stabilized, surface-attached.
- Tubulin: Green- and red-labelled GTP-tubulin.
- Tau Protein: Human 2N4R tau.
Workflow Summary:
- Grow Microtubules: Grow dynamic microtubules from stabilized seeds using green-labelled GTP-tubulin.
- Cap Microtubules: Inhibit further tip dynamics by capping with GMPCPP-tubulin.
- Incubate with Tau and Tubulin: Incubate capped microtubules with red-labelled GTP-tubulin in the presence of Tau (e.g., 0 nM, 0.5 nM, 20 nM) for a set duration (e.g., 15-30 min).
- Wash and Image: Wash out free tubulin to reduce background and image the microtubules to visualize incorporated red tubulin.
- Quantify Incorporation: Measure the length and spatial frequency of red tubulin incorporation stretches. Compare conditions with and without Tau to quantify its effect on lattice incorporation [4].
Assaying Tau-Mediated Tubulin Incorporation
Troubleshooting Common Experimental Challenges
FAQ 1: My microtubule preparations show unexpected structural heterogeneity or instability. What could be the cause?
Unexpected lattice heterogeneity is not necessarily an artifact; it is an intrinsic property of microtubules. "Holes" or multi-seam structures can form naturally within the shaft, acting as sites for tubulin exchange [82] [80].
- Solution: Implement structural analysis techniques like Segmented Subtomogram Averaging (SSTA) to distinguish between normal intrinsic heterogeneity and experimental artifacts [82]. Ensure that specimen denaturation at air-water interfaces during cryo-EM preparation is minimized [82].
FAQ 2: I am observing high background noise in my tubulin incorporation assays, making quantification difficult.
High background fluorescence is a common issue that obscures the signal from incorporated tubulin.
- Solution: After the incubation step with fluorescently labelled tubulin, perform a thorough wash with a tubulin-free buffer before imaging. This critical step reduces background signal and allows for clearer visualization and more accurate quantification of incorporated tubulin [4].
FAQ 3: My MTA treatment yields variable results between cell lines and in vitro assays. How can I improve validation?
Variable responses can stem from differences in tubulin isotype expression, efflux pumps, or the presence of specific MAPs across models.
- Solution: Emphasize cross-species validation. Integrate data from human tissues, animal models (e.g., mouse tauopathy models), and complementary in vivo systems (e.g., Drosophila) to prioritize high-confidence therapeutic targets and confirm conserved mechanisms of action [84]. This approach helps distinguish core drug effects from model-specific artifacts.
FAQ 4: Are there strategies to overcome the neurotoxicity and drug resistance associated with classic MTAs?
Yes, current research focuses on developing agents with novel binding sites and improved properties.
- Solution:
- Target New Sites: Investigate compounds binding to novel sites on tubulin, such as the gatorbulin site, which may offer alternative mechanisms to circumvent resistance [81] [39].
- Explore Selective Destabilizers: Develop compounds like benzimidazoles that show selectivity for specific β-tubulin isotypes (e.g., βVI), potentially reducing off-target effects in non-cancerous cells [81].
- Utilize Advanced Modalities: Consider Antibody-Drug Conjugates (ADCs) using agents like maytansine or dolastatin to improve tumor-specific targeting and reduce systemic toxicity [81].
Table 2: Key Research Reagent Solutions for Microtubule Studies
| Reagent / Resource | Function / Application | Key Details / Examples |
|---|---|---|
| Kinesin Motor-Domains | Decorates microtubules for structural studies; binds every αβ-tubulin heterodimer to reveal lattice organization [82] [83]. | Essential for SSTA to determine protofilament arrangement and seam locations [82]. |
| GMPCPP | Non-hydrolysable GTP analog; used to form stable microtubule "seeds" and cap dynamic ends [4]. | Stabilizes microtubules for controlled polymerization assays and lattice exchange experiments [4]. |
| Tau Protein | Key Microtubule-Associated Protein (MAP) studied for its role in lattice remodeling and stabilization [4]. | Human 2N4R isoform accelerates tubulin exchange at defect sites, challenging its view as a purely passive stabilizer [4]. |
| Software: IMOD & PEET | Standard software packages for processing electron tomograms and performing subtomogram averaging [82] [83]. | Used for 3D modeling, subvolume extraction, and averaging to achieve high-resolution structural insights [82]. |
| Novel Tubulin Binders | Tools for probing different binding sites and mechanisms to overcome resistance. | Gatorbulin-1 (binds a new site) [81] [39]; Pironetin (only known pure α-tubulin ligand) [81] [79]. |
Conclusion
The paradigm of microtubule lattice dynamics has fundamentally shifted from a static structure to a highly dynamic and heterogeneous entity central to cellular function. Key takeaways include the critical role of structural defects like seams and vacancies in governing lattice turnover and mechanical integrity, the sophisticated methods now available to characterize these phenomena, and the profound influence of MAPs like tau and pharmacological agents on lattice properties. The validation of microtubule instability as an early event in neurodegeneration opens new avenues for diagnostic biomarkers, while refined assays for drug evaluation promise more predictive development of microtubule-targeting therapies. Future research must focus on elucidating the precise regulation of lattice dynamics in vivo, developing next-generation imaging tools for clinical application, and designing novel therapeutic strategies that specifically modulate lattice repair and resilience, ultimately bridging fundamental biophysical insights with transformative clinical applications.
References
- 1. Lattice Structure of Cytoplasmic Microtubules in a Cultured Mammalian Cell - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2784118/]
- 2. Direct visualization of the microtubule lattice seam both in vitro and in vivo - PubMed [https://pubmed.ncbi.nlm.nih.gov/7806574/]
- 3. Competition for microtubule lattice spacing between a ... [https://www.sciencedirect.com/science/article/abs/pii/S0960982225010255]
- 4. Tau accelerates tubulin exchange in the microtubule lattice [https://www.nature.com/articles/s41567-025-03003-7]
- 5. A New Protocol to Accurately Determine Microtubule Lattice ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4634897/]
- 6. Ectopic A-lattice seams destabilize microtubules [https://www.nature.com/articles/ncomms4094]
- 7. [2511.06879] Beyond the Tip: Lattice Dynamics, Seams ... [https://arxiv.org/abs/2511.06879]
- 8. Double Seam Can Inspection: Frequently Asked Questions [https://industrialphysics.com/knowledgebase/articles/can-seam-inspection-frequently-asked-questions/]
- 9. Lattice defects induce microtubule self-renewal [https://www.hfsp.org/hfsp-news/lattice-defects-induce-microtubule-self-renewal]
- 10. Beyond the Tip: Lattice Dynamics, Seams, and ... [https://arxiv.org/html/2511.06879v1]
- 11. Tau accelerates tubulin exchange in the microtubule lattice [https://biophys.uni-saarland.de/publication/103-natphys-2025/]
- 12. Tau accelerates tubulin exchange in the microtubule lattice [https://sciencecast.org/casts/cgnk4as1mtle]
- 13. Interface-acting nucleotide controls polymerization ... [https://elifesciences.org/reviewed-preprints/89231]
- 14. Structural switching of tubulin in the microtubule lattice - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12186500/]
- 15. Interface-acting nucleotide controls polymerization ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10945504/]
- 16. GDP-to-GTP exchange on the microtubule end can ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5221584/]
- 17. Self-repair protects microtubules from their destruction by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7611741/]
- 18. of Characterization Heterogeneity by Segmented... Microtubule Lattice [https://bio-protocol.org/en/bpdetail?id=4723&type=0]
- 19. Characterization of Microtubule Lattice Heterogeneity by ... [https://pubmed.ncbi.nlm.nih.gov/37497446/]
- 20. Characterization of Microtubule Lattice Heterogeneity by ... [https://en.bio-protocol.org/en/bpdetail?id=4723&type=0]
- 21. Changes in seam number and location induce holes within ... [https://elifesciences.org/articles/83021]
- 22. A Practical Guide to Surface Kinetic Monte Carlo Simulations [https://pmc.ncbi.nlm.nih.gov/articles/PMC6465329/]
- 23. The Kinetic Monte Carlo method [https://www.sciencedirect.com/science/article/abs/pii/S0045782508001254]
- 24. Nanoscale characterization of drug-induced microtubule ... [https://pubmed.ncbi.nlm.nih.gov/34895240/]
- 25. Molecular probes for super-resolution imaging of drug ... [https://www.sciencedirect.com/science/article/abs/pii/S0169409X24001522]
- 26. A guide to choosing the right super-resolution microscopy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8246591/]
- 27. ELISA Tips: Troubleshooting Common Challenges - Blog [https://www.arp1.com/blog/post/elisa-kit-troubleshooting-tips.html]
- 28. Troubleshooting Immunoassays [https://www.anshlabs.com/resources/immunoassay-troubleshooting/]
- 29. In Vitro Reconstitution and Imaging of Microtubule ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7757405/]
- 30. In Vitro Assays to Study Force Generation at Dynamic ... [https://www.sciencedirect.com/science/article/abs/pii/S0091679X10950310]
- 31. In vitro reconstitution of dynamically interacting integral ... [https://elifesciences.org/articles/64389]
- 32. Microtubule Dynamics: an interplay of biochemistry and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6019280/]
- 33. Microtubule/Tubulin In Vivo Assay Biochem Kit [https://www.cytoskeleton.com/bk038]
- 34. In vitro Microtubule Binding Assay and Dissociation ... [https://en.bio-protocol.org/en/bpdetail?id=1759&type=0]
- 35. Real-Time Visualization of Microtubule and Protofilament ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10439732/]
- 36. - Microtubule : Advances in Tubulin Binding and... Targeting Agents [https://pubmed.ncbi.nlm.nih.gov/40806780/]
- 37. sciencedirect.com/topics/chemistry/ microtubule - stabilising - agent [https://www.sciencedirect.com/topics/chemistry/microtubule-stabilising-agent]
- 38. Diverse microtubule - targeted anticancer agents kill... | PLOS Biology [https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3002339]
- 39. Recent Advances in Microtubule Targeting Agents for ... [https://www.mdpi.com/1420-3049/30/16/3314]
- 40. Drugs That Target Dynamic Microtubules: A New Molecular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3155728/]
- 41. Microtubule-Targeting Agents: Advances in Tubulin ... [https://www.mdpi.com/1422-0067/26/15/7652]
- 42. Inhibition of p38-MK2 pathway enhances the efficacy ... [https://elifesciences.org/articles/104859]
- 43. Pharmacology and pharmacokinetics of antibody-drug ... [https://www.sciencedirect.com/science/article/abs/pii/S0305737225000441]
- 44. Imaging microtubule dynamics: A new frontier in biomarker ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12455249/]
- 45. Tau: Not Just a Stabilizer, But an Active Microtubule Remodeler [https://liphy.univ-grenoble-alpes.fr/en/news/tau-not-just-stabilizer-active-microtubule-remodeler]
- 46. Tau stabilizes microtubules by binding at the interface ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4475932/]
- 47. Graded regulation of microtubule-binding of Tau by the ... [https://pubmed.ncbi.nlm.nih.gov/41265727/]
- 48. Role of Tau as a Microtubule-Associated Protein: Structural ... [https://www.frontiersin.org/journals/aging-neuroscience/articles/10.3389/fnagi.2019.00204/full]
- 49. Tau oligomerization on microtubules in health and disease [https://pmc.ncbi.nlm.nih.gov/articles/PMC10841430/]
- 50. Microtubule Mysteries Revealed [https://tacc.utexas.edu/news/latest-news/2025/04/02/microtubule-mysteries-revealed/]
- 51. Developing Robust In Vitro Cell-Based Assays for High ... [https://www.marinbio.com/developing-robust-in-vitro-cell-based-assays-for-high-throughput-screening-hts/]
- 52. Top 5 Drug Discovery Trends 2025 Driving Breakthroughs [https://www.pelagobio.com/cetsa-drug-discovery-resources/blog/drug-discovery-trends-2025/]
- 53. Advanced In Vitro Models for Preclinical Drug Safety [https://pmc.ncbi.nlm.nih.gov/articles/PMC11763796/]
- 54. ADME Optimisation 2025 | Accelerate Drug Development [https://www.pharmaron.com/pharmaron-stories/adme-optimisation-2025/]
- 55. TIPsy tour guides: how microtubule plus-end tracking proteins ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4485311/]
- 56. Resolving bundled microtubules using anti-tubulin ... [https://www.nature.com/articles/ncomms8933]
- 57. Accessibility of the unstructured α-tubulin C-terminal tail is ... [https://elifesciences.org/reviewed-preprints/109308]
- 58. Protocol for lattice light-sheet time-lapse imaging of early post ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12593599/]
- 59. High-fidelity tissue super-resolution imaging achieved with ... [https://www.nature.com/articles/s41377-025-01930-x]
- 60. Super-oscillatory lattices structured illumination microscopy ... [https://www.sciencedirect.com/science/article/abs/pii/S0143816623004414]
- 61. High resolution microtubule structures reveal the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4054694/]
- 62. Discovery of novel microtubule destabilizing agents via ... [https://www.sciencedirect.com/science/article/abs/pii/S0169260725004146]
- 63. the diverse effects of microtubule-targeting drugs on tumor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12081712/]
- 64. Microtubule-Targeting Agents: Advances in Tubulin Binding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12347508/]
- 65. Association of microtubule destabilization with platelet yields ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0326165]
- 66. Spatial Statistics of Three-Dimensional Growth Dynamics ... [https://pubmed.ncbi.nlm.nih.gov/39616568/]
- 67. Polyglutamylation of microtubules drives neuronal ... [https://www.nature.com/articles/s41467-025-60855-6]
- 68. Tubulin Polymerization Assay - an overview [https://www.sciencedirect.com/topics/medicine-and-dentistry/tubulin-polymerization-assay]
- 69. Tubulin and microtubule based assays, compound ... [https://www.cytoskeleton.com/custom-services3-compound-screening-tubulin-microtubules]
- 70. Establishing a High-content Analysis Method for Tubulin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3941064/]
- 71. Measurement of In Vitro Microtubule Polymerization by ... [https://www.sciencedirect.com/science/chapter/bookseries/abs/pii/B9780124077577000141]
- 72. In vitro tubulin polymerization assay [https://bio-protocol.org/exchange/minidetail?id=17420567&type=30]
- 73. Cell Imaging Support | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/cell-analysis-support-center/cell-imaging-support.html]
- 74. A cell-level model to predict the spatiotemporal dynamics ... [https://arxiv.org/html/2508.15046v1]
- 75. Integrating digital pathology and mathematical modelling to ... [https://www.nature.com/articles/s41746-022-00636-3]
- 76. Biomarkers and therapeutic strategies targeting microglia in ... [https://molecularneurodegeneration.biomedcentral.com/articles/10.1186/s13024-025-00867-4]
- 77. Preliminary mechanistic insights of a brain-penetrant ... [https://ejnmmires.springeropen.com/articles/10.1186/s13550-022-00912-z]
- 78. Binding Parameters of [ 11 C]MPC-6827, a Microtubule- ... [https://www.mdpi.com/1424-8247/16/4/495]
- 79. Recent Approaches to the Identification of Novel ... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2022.841777/full]
- 80. Beyond uniformity: Exploring the heterogeneous and ... [https://www.sciencedirect.com/science/article/pii/S0171933523000857]
- 81. The microtubule cytoskeleton: An old validated target for novel ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9521402/]
- 82. Characterization of Microtubule Lattice Heterogeneity by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10366996/]
- 83. Characterization of Microtubule Lattice Heterogeneity by ... [https://bio-protocol.org/pdf/Bio-protocol4723.pdf]
- 84. Cross species systems biology discovers glial DDR2 ... [https://www.nature.com/articles/s41467-023-42626-3]