Mastering Actin Fiber Analysis: A Complete SFEX Software Tutorial for Cell Biology Research
This comprehensive tutorial provides researchers and drug development professionals with a complete guide to SFEX (Stress Fiber Extractor) software for accurate actin fiber segmentation and quantification.
Mastering Actin Fiber Analysis: A Complete SFEX Software Tutorial for Cell Biology Research
Abstract
This comprehensive tutorial provides researchers and drug development professionals with a complete guide to SFEX (Stress Fiber Extractor) software for accurate actin fiber segmentation and quantification. The article covers foundational concepts of stress fiber biology and SFEX's role, a step-by-step methodological walkthrough from installation to batch processing, expert troubleshooting and optimization techniques for challenging images, and validation protocols comparing SFEX to manual and other automated methods. The guide enables reproducible, high-throughput analysis of cytoskeletal morphology for applications in cell mechanics, disease modeling, and drug discovery.
Understanding Stress Fibers and SFEX: Core Concepts for Quantitative Cytoskeleton Analysis
Why Actin Fiber Quantification is Crucial in Biomedical Research
Actin fibers, or microfilaments, are fundamental components of the cytoskeleton, governing cell morphology, migration, division, and mechanotransduction. Precise quantification of actin fiber properties—such as density, orientation, length, and thickness—transcends simple descriptive microscopy, offering critical, quantitative insights into cellular state and function. Within biomedical research, this quantification is indispensable for elucidating disease mechanisms, screening drug efficacy, and understanding fundamental cell biology. The advent of automated, robust software tools like SFEX (Stress Fiber Extractor) has revolutionized this field by enabling high-throughput, reproducible analysis from standard fluorescence images, moving beyond subjective qualitative assessment.
The Critical Role in Research and Drug Development
Quantitative actin analysis provides actionable data across multiple domains:
- Cancer Research: Correlating actin fiber organization with metastatic potential and invasiveness. More aligned, contractile stress fibers are often associated with increased migratory capacity.
- Cardiovascular Disease: Assessing cardiomyocyte health and sarcomeric organization, where actin disarray is a hallmark of conditions like hypertrophic cardiomyopathy.
- Neurodegenerative Disorders: Investigating the role of the actin cytoskeleton in growth cone dynamics, synaptic stability, and pathological aggregate formation.
- Drug Discovery & Toxicology: Using cytoskeletal remodeling as a sensitive phenotypic endpoint for screening compound libraries or assessing off-target toxic effects. Drugs affecting Rho GTPase pathways, for instance, produce distinct, quantifiable actin phenotypes.
- Mechanobiology: Quantifying cellular responses to substrate stiffness by measuring stress fiber assembly and alignment.
The transition to software-based quantification addresses key challenges: eliminating observer bias, enabling analysis of large datasets (e.g., from high-content screening), and extracting subtle phenotypic differences invisible to the human eye.
Application Notes: From Image to Insight with SFEX
The following protocol outlines a standard workflow for quantifying actin fibers in cultured cells, such as vascular smooth muscle cells (VSMCs) stimulated to form stress fibers, using SFEX software within a broader research thesis context.
Table 1: Key Quantitative Outputs from Actin Fiber Analysis
| Quantitative Metric | Biological Interpretation | Example Application |
|---|---|---|
| Fiber Density | Total polymerized actin per cell/area. | Measuring cytoskeletal collapse upon toxin exposure. |
| Alignment Index | Degree of directional order (0=random, 1=perfectly aligned). | Assessing fibroblast polarization during wound healing. |
| Average Fiber Length | Stability and polymerization dynamics of filaments. | Evaluating the effect of actin-stabilizing drugs. |
| Orientation Angle | Preferred directional bias of fibers relative to a reference. | Studying contact guidance on micro-patterned surfaces. |
Detailed Experimental Protocol
Part A: Cell Culture, Stimulation, and Immunofluorescence
This protocol generates samples with defined actin phenotypes suitable for quantitative analysis.
I. Materials & Reagents (Research Reagent Solutions) Table 2: Essential Materials for Actin Staining and Imaging
| Item | Function / Description |
|---|---|
| Vascular Smooth Muscle Cells (A7r5 line) | Model system for inducible stress fiber formation. |
| DMEM, 10% FBS, 1% P/S | Standard cell culture medium for maintenance. |
| Serum-Free DMEM | Medium for serum-starvation to induce a quiescent state. |
| Lysophosphatidic Acid (LPA, 10 µM stock) | Agonist that activates RhoA signaling to induce robust stress fiber formation. |
| 4% Paraformaldehyde (PFA) | Fixative to preserve cellular architecture. |
| 0.1% Triton X-100 in PBS | Permeabilization agent for antibody access. |
| Phalloidin (e.g., Alexa Fluor 488 conjugate) | High-affinity probe that selectively stains filamentous (F-) actin. |
| Antibody Dilution Buffer (1% BSA in PBS) | Reduces non-specific antibody binding. |
| Microscope Slides & Coverslips (#1.5) | High-quality imaging substrates. |
| Mounting Medium with DAPI | Preserves fluorescence and stains nuclei for segmentation. |
| Inverted Epifluorescence or Confocal Microscope | Equipment for high-resolution image acquisition. |
II. Step-by-Step Procedure
- Culture & Plate Cells: Maintain A7r5 cells in complete growth medium. Plate cells at an appropriate density (e.g., 10,000 cells/cm²) onto sterile coverslips in a multi-well plate and culture overnight.
- Serum Starvation: Replace medium with serum-free DMEM for 24 hours to induce quiescence and reduce baseline stress fibers.
- Stimulation: Treat cells with 1-5 µM LPA (diluted from stock in serum-free DMEM) for 30-60 minutes. Include control wells with serum-free medium only.
- Fixation: Aspirate medium. Rinse gently with warm PBS. Fix cells with 4% PFA for 15 minutes at room temperature (RT). Rinse 3x with PBS.
- Permeabilization: Incubate cells with 0.1% Triton X-100 in PBS for 5-10 minutes at RT. Rinse 3x with PBS.
- Staining: Prepare phalloidin conjugate (e.g., 1:200) in antibody dilution buffer. Apply to coverslips and incubate for 60 minutes at RT in the dark.
- Nuclear Counterstain: Rinse 3x with PBS. Apply a drop of mounting medium with DAPI and mount the coverslip onto a microscope slide.
- Image Acquisition: Using a 40x or 60x oil-immersion objective, acquire Z-stack or single-plane images of the actin channel (e.g., FITC/488 nm for Alexa Fluor 488 phalloidin) and the DAPI channel. Maintain consistent exposure settings across all experimental conditions.
Part B: Quantitative Analysis with SFEX Software
This protocol details the segmentation and quantification process.
- Software Launch & Import: Open SFEX. Import your actin channel image (TIFF format recommended). Ensure the image scale (µm/pixel) is correctly set in the software settings.
- Pre-processing: Apply necessary pre-processing steps within SFEX: a) Subtract background fluorescence. b) Use a mild Gaussian blur filter to reduce noise, if required.
- Fiber Segmentation: Execute the core SFEX algorithm. This typically involves: a) Enhanced Ridge Detection to identify linear fiber structures. b) Adaptive Thresholding to distinguish fibers from background. c) Skeletonization to reduce fibers to single-pixel-width representations for measurement.
- Post-processing & Filtering: Use built-in filters to remove objects below a minimum length (e.g., <1 µm) to exclude noise and small fragments.
- Quantification: Run the analysis module to compute metrics for each cell or field of view. Key outputs include: Fiber Density (total fiber length/area), Alignment Index, and Average Fiber Length.
- Data Export: Export the numerical data table (e.g., to .csv format) for statistical analysis in external software (e.g., GraphPad Prism, R).
Signaling Pathways and Workflow Visualization
LPA-Induced Actin Polymerization Pathway
Actin Quantification Experimental Workflow
Application Notes and Protocols
Within the broader framework of a thesis on utilizing the SFEX (Stress Fiber Extractor) software for actin fiber segmentation research, these notes provide foundational context. SFEX is a critical tool for quantifying cytoskeletal reorganization, a phenotypic marker in mechanobiology, toxicology, and drug discovery.
Development History
SFEX was developed to address the lack of automated, quantitative tools for analyzing stress fibers from fluorescence microscopy images. Its evolution is marked by key algorithmic improvements.
| Version / Milestone | Year | Key Advancement | Impact on Actin Research |
|---|---|---|---|
| Initial Concept | ~2014 | Manual preprocessing with basic ridge detection. | Demonstrated need for automation in fiber quantification. |
| SFEX v1.0 | 2016 | Introduction of core multi-scale Hessian-based ridge detection pipeline. | Enabled first large-scale, reproducible segmentation of fibers. |
| SFEX with GUI (v2.0) | 2018 | Integration of a graphical user interface and batch processing. | Increased accessibility for biologists, allowing high-throughput analysis. |
| SFEX-AI (Current) | 2022+ | Integration of deep learning modules (e.g., U-Net) for enhanced segmentation of dense or complex networks. | Improved accuracy in challenging conditions, such as confluent cells or in vivo tissues. |
Core Algorithm Principles
The core of SFEX is a multi-scale, Hessian matrix-based ridge detection algorithm optimized for linear structures.
Core Workflow Protocol
Protocol: Image Processing for Stress Fiber Extraction using SFEX Core Algorithm
Input: 2D fluorescent image of actin (e.g., phalloidin stain). Output: Binary mask of detected stress fibers and quantitative metrics (orientation, length, width, alignment).
Steps:
- Preprocessing: Apply Gaussian blur (σ=1-2 pixels) to reduce high-frequency noise.
- Hessian Matrix Calculation: For each pixel at scale σ, compute the second-order partial derivatives (Ixx, Iyy, Ixy) to form the Hessian matrix H.
- H = [[Ixx, Ixy], [Ixy, Iyy]]
- Eigenvalue Analysis: Calculate eigenvalues (λ1, λ2, where |λ1| ≥ |λ2|) and eigenvectors of H.
- The eigenvector corresponding to λ1 indicates the ridge direction perpendicular to the fiber.
- Vesselness / Ridgeness Measurement: Apply a fiber-enhanced likelihood function. A common metric is derived from Frangi's vesselness filter:
- V(σ) = 0 if λ1 > 0,
- V(σ) = exp(-Rβ²/2β²) * (1 - exp(-S²/2c²)) otherwise.
- Rβ = |λ2|/|λ1| (differentiates line from blob).
- S = sqrt(λ1² + λ2²) (differentiates structure from background).
- β, c are sensitivity parameters.
- Multi-scale Integration: Repeat steps 2-4 across a defined range of scales (e.g., σ = 1-5 pixels). For each pixel, retain the maximum V(σ) value across scales.
- Post-processing: Apply adaptive thresholding (Otsu's method) on the integrated vesselness map to create a binary mask. Optional morphological cleaning (spur removal) may be applied.
SFEX Core Algorithm Workflow
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in Actin/SFEX Research |
|---|---|
| Fluorescent Phalloidin (e.g., Alexa Fluor 488, 568, 647 conjugates) | High-affinity F-actin probe for staining stress fibers in fixed cells for SFEX analysis. |
| Live-Actin Probes (e.g., LifeAct-EGFP, SiR-actin) | Enables live-cell imaging of actin dynamics; SFEX can analyze time-series data. |
| Rho GTPase Modulators (e.g., Lysophosphatidic Acid - LPA, Y-27632 ROCK inhibitor) | Pharmacological tools to induce or disrupt stress fibers, validating SFEX's sensitivity to phenotypic change. |
| Matrigel / Stiffness-Tunable Hydrogels (e.g., Polyacrylamide gels) | Substrates to study mechanotransduction. SFEX quantifies how substrate stiffness influences fiber density and alignment. |
| High-Resolution Microscope (Confocal, TIRF, or Super-resolution) | Provides input images. Image quality (SNR, resolution) is the primary determinant of SFEX segmentation accuracy. |
| SFEX Software | The core analytical tool for automated, quantitative extraction of fiber morphology and orientation data. |
Experimental Validation Protocol
Protocol: Validating SFEX with Pharmacological Cytoskeletal Disruption
Aim: To demonstrate SFEX's ability to quantify drug-induced changes in actin cytoskeleton.
Materials:
- HeLa or NIH/3T3 cells.
- Standard cell culture materials.
- Lysophosphatidic Acid (LPA, 10 µM stock), Y-27632 (10 mM stock).
- Fluorescent phalloidin (as per Toolkit).
- 4% Paraformaldehyde (PFA), Triton X-100.
- Confocal microscope.
- SFEX software installed.
Methodology:
- Cell Seeding: Seed cells on glass coverslips in 24-well plates. Culture until ~70% confluency.
- Compound Treatment:
- Group 1 (Control): Serum-free medium for 16h, then fresh serum-free medium for 30 min.
- Group 2 (Fiber Inducer): Serum-starve 16h, then treat with 5 µM LPA (diluted in serum-free medium) for 30 min.
- Group 3 (Fiber Disruptor): Serum-starve 16h, treat with LPA (5 µM, 30 min), then add Y-27632 (10 µM) for 60 min.
- Fixation and Staining: Wash cells with PBS. Fix with 4% PFA for 15 min. Permeabilize with 0.1% Triton X-100 for 5 min. Stain with fluorescent phalloidin (1:500 in PBS) for 30 min in the dark. Mount slides.
- Imaging: Acquire 5-10 representative 60x images per condition using constant exposure settings.
- SFEX Analysis: Process all images through SFEX using identical parameters (scale range: 1-5, default β, c). Export fiber area density and average alignment index.
- Statistical Analysis: Perform one-way ANOVA with post-hoc test on the quantitative outputs from SFEX (n≥30 images total).
SFEX Validation Experimental Workflow
Application Note 1: Quantifying Cytoskeletal Remodeling in Response to Substrate Stiffness
Context: A core thesis chapter demonstrates SFEX's ability to quantify actin architecture changes, a fundamental readout of cellular mechanotransduction. This protocol details its application in a classic cell mechanics experiment.
Protocol: Actin Fiber Analysis on Polyacrylamide Gels of Varying Stiffness
- Substrate Preparation: Fabricate polyacrylamide hydrogels with elastic moduli of 1 kPa (soft), 8 kPa (intermediate), and 25 kPa (stiff) using published protocols. Functionalize surfaces with 0.1 mg/mL collagen I.
- Cell Culture & Plating: Seed NIH/3T3 fibroblasts at 5,000 cells/cm² on each gel condition. Allow cells to adhere and spread for 18 hours in complete DMEM.
- Fixation & Staining: Fix cells with 4% paraformaldehyde for 15 min, permeabilize with 0.1% Triton X-100, and stain actin filaments with Alexa Fluor 488-phalloidin (1:200) for 1 hour.
- Image Acquisition: Acquire 10 high-resolution (63x/1.4 NA oil objective) images per condition using a confocal microscope with identical laser power and gain settings.
- SFEX Segmentation & Analysis:
- Import images into SFEX Stress Fiber Extractor.
- Apply the "Standard Fiber Detection" pipeline with a contrast threshold of 0.15 and a minimum fiber length of 5 µm.
- Run batch analysis on all images from the same condition.
- Export quantitative metrics: Fiber Density (%), Mean Fiber Length (µm), and Alignment Index (0-1).
- Data Interpretation: Compare metrics across stiffness conditions to quantify the shift from isotropic networks (soft) to prominent, aligned stress fibers (stiff).
Quantitative Data Summary: Actin Architecture vs. Substrate Stiffness
| Substrate Stiffness (Elastic Modulus) | Mean Fiber Density (% of Cell Area) ± SD | Mean Fiber Length (µm) ± SD | Mean Alignment Index ± SD |
|---|---|---|---|
| 1 kPa (Soft) | 12.3 ± 2.1 | 7.8 ± 1.5 | 0.21 ± 0.08 |
| 8 kPa (Intermediate) | 24.7 ± 3.5 | 12.4 ± 2.2 | 0.45 ± 0.10 |
| 25 kPa (Stiff) | 38.9 ± 4.8 | 18.9 ± 3.1 | 0.72 ± 0.09 |
Table 1: SFEX-derived metrics show increased actin polymerization, bundling, and alignment with increasing substrate stiffness (n=50 cells per condition).
The Scientist's Toolkit: Research Reagent Solutions
| Item/Reagent | Function in Experiment |
|---|---|
| Polyacrylamide Hydrogel Kit | Provides tunable, physiologically relevant substrate stiffness for cell culture. |
| Collagen I, Bovine | ECM protein coating for gel functionalization to enable integrin-mediated cell adhesion. |
| Alexa Fluor 488-phalloidin | High-affinity, fluorescent probe for selective staining of filamentous actin (F-actin). |
| Paraformaldehyde (4%) | Crosslinking fixative that preserves cytoskeletal architecture. |
| Triton X-100 | Non-ionic detergent for cell permeabilization, allowing phalloidin entry. |
Experimental Workflow: Substrate Stiffness Assay
Application Note 2: High-Content Screening for Anti-Fibrotic Drug Candidates
Context: This application note, relevant to a thesis on SFEX's scalability, outlines its use in a drug discovery pipeline to identify compounds that disrupt pathological stress fiber formation.
Protocol: High-Content Screening for Actin De-polymerizers
- Cell Model & Induction: Seed TGF-β1-sensitive human lung fibroblasts (HLF-1) in 384-well imaging plates. Treat with 5 ng/mL TGF-β1 for 48 hours to induce a myofibroblast phenotype with robust stress fibers.
- Compound Library Treatment: Co-treat cells with TGF-β1 and a library of small molecule compounds (10 µM each) or DMSO control. Include positive control (e.g., 5 µM Latrunculin B).
- Automated Staining & Imaging: After 48h, automate fixation and staining using Hoechst 33342 (nuclei) and Alexa Fluor 555-phalloidin (actin). Acquire 9 fields/well using a 20x objective on a high-content imaging system.
- Automated SFEX Analysis Pipeline:
- Use the SFEX API or batch scripting to process all fields.
- Apply a consistent segmentation threshold. Use the "Cell Profiler Integration" module to link actin metrics to individual nuclei.
- Key output metric: Mean Fiber Density per Cell.
- Hit Identification & Validation: Normalize data to DMSO (TGF-β1 only = 100%) and Latrunculin B (0%). Primary hits: compounds reducing fiber density >40%. Confirm hits in dose-response and secondary assays (e.g., α-SMA expression).
Quantitative Data Summary: Sample Screening Results
| Treatment Condition (with TGF-β1) | Mean Fiber Density (% of Control) ± SEM | Z'-Factor (Plate-Wise) | Hit Classification |
|---|---|---|---|
| DMSO (Vehicle Control) | 100.0 ± 3.2 | 0.62 | N/A |
| Latrunculin B (5 µM) | 18.5 ± 2.1 | N/A | Positive Control |
| Compound A | 92.4 ± 4.5 | N/A | Inactive |
| Compound B | 45.7 ± 3.8 | N/A | Primary Hit |
| Compound C | 32.1 ± 2.9 | N/A | Primary Hit |
Table 2: SFEX-enabled HCS identifies compounds B and C as potent disruptors of TGF-β1-induced stress fiber formation. Z'-factor indicates a robust assay.
Pathway Diagram: TGF-β / Actin Signaling in Fibrosis
The Scientist's Toolkit: HCS Essentials
| Item/Reagent | Function in Experiment |
|---|---|
| TGF-β1, Human Recombinant | Cytokine to induce myofibroblast differentiation and stress fiber formation. |
| 384-Well Glass-Bottom Plates | Optically clear plates suitable for automated, high-resolution microscopy. |
| Small Molecule Library | Diverse chemical collection for primary screening of actin modulators. |
| Latrunculin B | Actin de-polymerizing toxin used as a robust positive control in the assay. |
| Hoechst 33342 | Cell-permeable nuclear stain for automated cell segmentation and counting. |
| Automated Liquid Handler | Enables reproducible compound dispensing and staining for high-throughput workflows. |
System Requirements and Software Installation Guide (Windows/macOS/Linux)
This guide details the installation and configuration of the SFEX (Stress Fiber Extractor) software, a critical tool for the quantitative analysis of actin cytoskeleton organization in biomedical research. Proper installation is a prerequisite for the high-throughput segmentation and quantification of stress fibers from fluorescence microscopy images, enabling studies in cell mechanics, drug response, and disease modeling.
System Requirements
The following tables summarize the minimum and recommended hardware and software requirements for SFEX across supported operating systems. SFEX is distributed as a platform-specific installer or as a Python package.
Table 1: Hardware Requirements
| Component | Minimum Requirements | Recommended Specifications |
|---|---|---|
| CPU | 64-bit, 2 cores | 64-bit, 8+ cores |
| RAM | 8 GB | 16 GB or more |
| Storage | 1 GB free space (SSD recommended) | 10+ GB free space (NVMe SSD) |
| GPU | Integrated graphics | Dedicated GPU (NVIDIA with CUDA support for accelerated processing) |
| Display | 1280x720 resolution | 1920x1080 resolution or higher |
Table 2: Software Requirements & Dependencies
| OS | SFEX Installer Version | Python Package Requirements | Mandatory System Dependencies |
|---|---|---|---|
| Windows | 10 or 11 (64-bit) | Python 3.8-3.10 | Microsoft Visual C++ Redistributable 2019 |
| macOS | 11 (Big Sur) or later | Python 3.8-3.10 | Xcode Command Line Tools |
| Linux | Ubuntu 20.04/22.04, CentOS 7/8 | Python 3.8-3.10 | GCC, libgl1-mesa-glx, libgtk2.0-0 |
Installation Protocols
Protocol 3.1: Installation via Official Installer (Windows/macOS)
Objective: To install SFEX and its core dependencies using the graphical installer for simplified setup.
- Download: Obtain the latest installer for your OS from the official SFEX repository (e.g.,
SFEX_Setup_Windows_v2.1.0.exeorSFEX_Mac_v2.1.0.pkg). - Run Installer:
- Windows: Double-click the
.exefile, grant administrator permissions if prompted, and follow the on-screen instructions. - macOS: Double-click the
.pkgfile. If blocked by Gatekeeper, right-click and select "Open."
- Windows: Double-click the
- Path Configuration: The installer will automatically add SFEX to your system's PATH. Verify installation by opening a new terminal/command prompt and typing
sfex --version.
Protocol 3.2: Installation via Python Package Manager (All OS)
Objective: To install SFEX within a controlled Python environment, ideal for integration into custom analysis pipelines.
- Create a Virtual Environment (Recommended):
Update Package Tools:
Install SFEX:
Verify Installation: Run
python -c "import sfex; print(sfex.__version__)".
Protocol 3.3: Installation from Source (Linux/macOS)
Objective: To install the latest development version for access to cutting-edge features.
- Clone Repository:
Install in Editable Mode:
Run Tests: Execute
pytestto confirm all components are functional.
Post-Installation Validation & Basic Workflow
Protocol 4.1: Software Validation Test
- Launch SFEX from the command line:
sfex-gui(for GUI) or use the Python API. - Load the provided sample image (
sample_actin.tif). - Navigate to the Process tab and click Run Standard Analysis.
- Successful execution will generate a results folder containing segmented fiber masks and quantitative metrics (e.g., fiber alignment, density, length).
Title: SFEX Actin Fiber Analysis Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Actin Fiber Imaging & SFEX Analysis
| Item | Function in Context of SFEX Analysis |
|---|---|
| Fluorescent Phalloidin (e.g., Alexa Fluor 488, 568, 647) | High-affinity stain for F-actin, providing the specific and high-contrast signal required for robust SFEX segmentation. |
| Cell Fixative (e.g., 4% Paraformaldehyde) | Preserves cellular architecture and actin cytoskeleton at the time of staining, preventing artifact generation. |
| Permeabilization Buffer (e.g., 0.1% Triton X-100) | Allows phalloidin to penetrate the cell membrane and bind to internal actin filaments. |
| Mounting Medium with Antifade | Preserves fluorescence signal during microscopy, preventing photobleaching that can degrade analysis quality. |
| High-NA Objective Lens (60x/100x oil immersion) | Essential for capturing high-resolution, detailed images of stress fibers that SFEX is designed to analyze. |
| SFEX Software & Compatible Python Environment | The core computational tool for converting raw image data into quantitative, statistically analyzable metrics. |
Troubleshooting Common Installation Issues
- "DLL load failed" on Windows: Install the latest Microsoft Visual C++ Redistributable.
- "Command not found: sfex" on macOS/Linux: Ensure the terminal shell is restarted or the virtual environment is activated.
- GUI fails to launch on Linux: Install required system libraries:
sudo apt-get install libgl1-mesa-glx libgtk2.0-0.
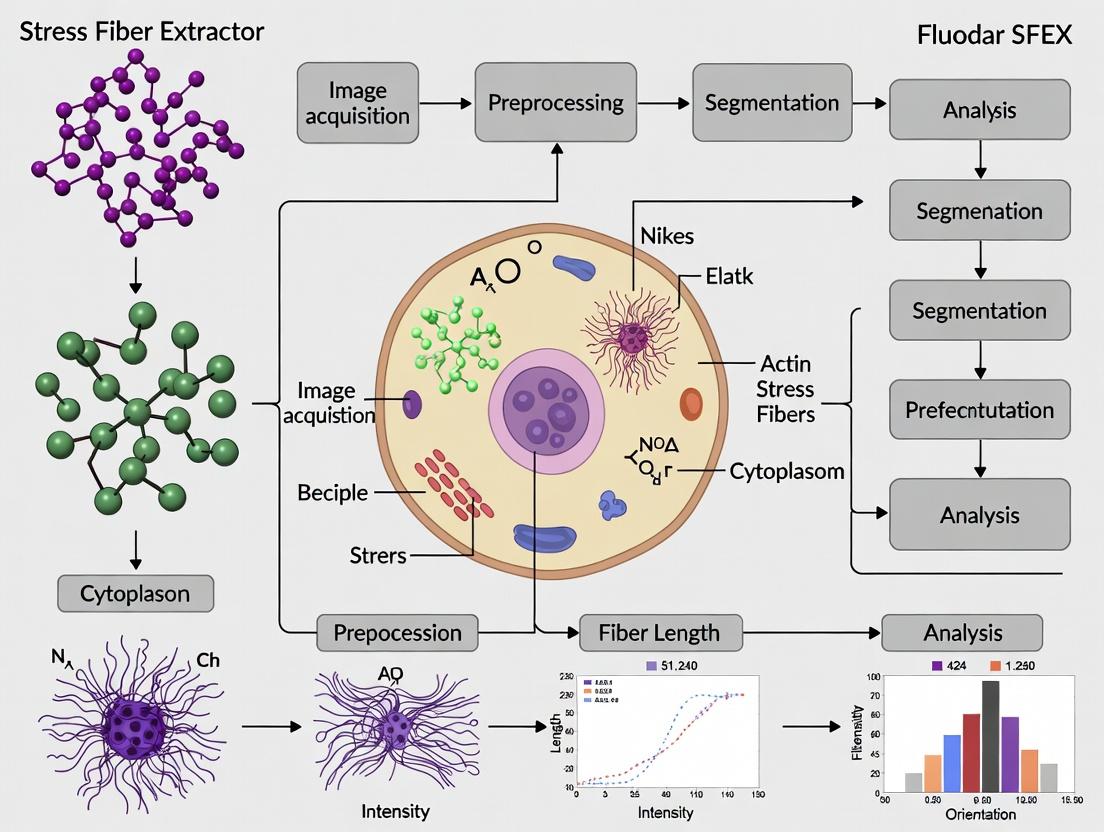
This application note provides an initial, detailed protocol for navigating the core user interface modules of the SFEX (Stress Fiber Extractor) software. Designed within the context of a comprehensive tutorial series for actin fiber segmentation in cellular research, this guide targets researchers, scientists, and drug development professionals. The note details the functional layout, data handling procedures, and essential workflows required to initiate quantitative analysis of stress fibers from fluorescence microscopy images, a critical step in phenotypic screening and mechanobiology studies.
SFEX is a specialized software tool for the automated extraction, segmentation, and quantitative analysis of actin stress fibers from 2D fluorescence micrographs. Accurate quantification of fiber morphology, alignment, and intensity is vital for research into cell mechanics, response to pharmacological agents, and disease states. This document focuses on the primary UI modules that form the foundation for all analytical workflows.
Key User Interface Modules & Functions
The main SFEX interface is divided into five core modules. The table below summarizes their primary functions and output data types.
Table 1: Core SFEX UI Modules and Functions
| Module Name | Primary Function | Key Outputs/Controls |
|---|---|---|
| Project Explorer | Manages raw image datasets, analysis parameters, and results hierarchies. | Directory tree, file lists, metadata viewer. |
| Image Viewer & Preprocessor | Displays raw and processed images; applies initial filters and contrast adjustments. | Z-projection tools, background subtraction, channel selector. |
| Segmentation Parameter Panel | Houses critical algorithms for fiber detection and binary mask creation. | Threshold sliders (Otsu, Li), fiber enhancement kernels, seed point controls. |
| Quantification Dashboard | Calculates and displays morphometric data from segmented fibers. | Data table for fiber length, width, alignment (cos²θ), intensity. |
| Visualization & Export | Generates overlays and plots; exports data for statistical analysis. | Fiber overlay on original image, rose plots, data export to CSV/TSV. |
Standard Protocol: Initial Image Analysis Workflow
This protocol outlines the essential steps from image loading to obtaining preliminary quantitative data.
Materials & Reagent Solutions
Table 2: Research Reagent Solutions for Actin Imaging
| Item | Function in Context |
|---|---|
| Fluorescent Phalloidin (e.g., Alexa Fluor 488, 555, 647) | High-affinity filamentous actin (F-actin) stain used to generate input images for SFEX. |
| Cell Fixative (e.g., 4% Paraformaldehyde in PBS) | Preserves cellular architecture and actin cytoskeleton at the time of assay. |
| Permeabilization Buffer (e.g., 0.1% Triton X-100) | Allows phalloidin to penetrate the cell membrane and bind to internal F-actin. |
| Mounting Medium with DAPI | Seals samples; DAPI stains nuclei for optional cell segmentation/co-localization. |
| Cultured Cells on Glass Coverslips | Standard substrate for high-resolution, flat imaging of the actin cytoskeleton. |
Step-by-Step Procedure
- Project Initialization: Launch SFEX. In the Project Explorer, select "New Project." Navigate to and select the directory containing your actin channel TIFF files (e.g., phalloidin stain). SFEX will catalogue all compatible images.
- Image Inspection & Pre-processing: Double-click an image in the Project Explorer to load it into the Image Viewer. Use the "Pre-process" sub-panel to:
- Apply a Z-projection (if dealing with a stack) using the Maximum Intensity method.
- Execute Background Subtraction using the Rolling Ball algorithm (radius: 50 pixels).
- Adjust Contrast/Limits manually to ensure clear fiber visibility.
- Segmentation Parameter Calibration: Navigate to the Segmentation Parameter Panel.
- Set Detection Method to "Hessian-based Enhancement."
- Adjust the Fiber Width parameter to match the approximate pixel width of fibers in your image (typical range: 3-7 px).
- Set the Threshold Method to "Otsu" and click "Calculate." Manually fine-tune the threshold slider if necessary.
- Click "Run Segmentation." The binary mask will appear in the Image Viewer.
- Quantification & Review: Upon segmentation, the Quantification Dashboard will auto-populate. Review key metrics for the sample:
- Fiber Density (total fiber area / total cell area)
- Mean Fiber Length
- Alignment Index (Directionality Order Parameter)
- Visualization & Export: Go to the Visualization & Export module.
- Select "Overlay Fibers" to superimpose detected fibers (in green) onto the original grayscale image.
- Generate a "Rose Plot" of fiber orientations.
- Select all data in the Quantification Dashboard and export to CSV format for external analysis.
Schematic Workflows
Title: SFEX Core Analysis Workflow
Title: SFEX Primary UI Module Relationships
Step-by-Step SFEX Workflow: From Image Import to Quantified Data Export
This application note details best practices for preparing microscopy images for use in SFEX Stress Fiber Extractor software, a tool for automated segmentation and quantification of actin stress fibers. Proper image preparation is critical for ensuring the accuracy and reproducibility of downstream analysis within actin cytoskeleton research and drug development workflows.
Image File Format Specifications
The choice of file format directly impacts data integrity. SFEX requires single-channel, 2D grayscale images or single-plane fluorescent images for optimal fiber extraction.
| Format | Key Characteristics | Best For SFEX? | Primary Rationale |
|---|---|---|---|
| TIFF (Tagged Image File Format) | Uncompressed or lossless compression (LZW). Supports 8, 16, 32-bit depth. Can store metadata. | Yes, Preferred | Preserves full dynamic range. No data loss. Common in microscopy. |
| PNG (Portable Network Graphics) | Lossless compression. Typically 8-bit depth (16-bit supported). No native metadata standard. | Yes, Acceptable | Good for 8-bit data. Smaller file size than uncompressed TIFF. |
| JPEG (Joint Photographic Experts Group) | Lossy compression. 8-bit depth. Artifacts can obscure fine fibers. | No | Compression artifacts degrade edge detection and segmentation. |
| ND2 (Nikon), LIF (Leica), CZI (Zeiss) | Proprietary, multi-dimensional formats (stack, time, channel). | No (Directly) | Must be exported as single-plane TIFF/PNG. Use manufacturer software or Bio-Formats. |
Image Acquisition & Pre-processing Protocol
Objective: To acquire and pre-process microscopy images of fluorescently labeled actin (e.g., with phalloidin) to maximize SFEX segmentation performance.
Materials & Reagents:
- Research Reagent Solutions:
- Fluorescent Phalloidin Conjugates: Binds filamentous actin (F-actin) with high specificity. Function: Target staining.
- Cell Fixative (e.g., 4% PFA): Preserves cellular architecture at time of fixation. Function: Sample fixation.
- Permeabilization Buffer (e.g., 0.1% Triton X-100): Allows dye penetration. Function: Membrane permeabilization.
- Mounting Medium with Anti-fade: Reduces photobleaching. Function: Slide preservation.
- High-NA Objective Lens (60x/100x): Essential for resolving individual fibers. Function: High-resolution imaging.
- Scientific CMOS (sCMOS) Camera: Provides high dynamic range and low noise. Function: Signal detection.
Protocol:
- Sample Preparation: Culture cells on glass-bottom dishes. Fix with 4% PFA for 15 min. Permeabilize with 0.1% Triton X-100 for 5 min. Stain with fluorescent phalloidin (e.g., Alexa Fluor 488, 1:200) for 30 min in the dark. Mount with anti-fade medium.
- Microscopy Acquisition:
- Use a 60x or higher magnification oil-immersion objective (NA ≥ 1.4).
- Set the camera gain to a constant, mid-range value.
- Critical: Adjust laser/power and exposure time to avoid pixel saturation. Use the histogram tool to ensure signal occupies the dynamic range without clipping at 0 or 4095 (for 12-bit) or 65535 (for 16-bit).
- Focus: Acquire a single, in-focus Z-plane containing the majority of stress fibers.
- Save: Acquire data in the microscope's native format (e.g., .nd2, .lif, .czi) to retain all metadata.
Image Export & Formatting Workflow for SFEX
Objective: To convert proprietary multi-dimensional image data into SFEX-compatible 2D grayscale images.
Protocol:
- Open & Select: Open the proprietary file in Fiji/ImageJ using the Bio-Formats Importer plugin.
- Split Channels: If multi-channel, split channels and retain only the actin (phalloidin) channel.
- Select Z-plane: If a stack, use the
Z-projectfunction (chooseMax Intensity) or manually select the single plane with the sharpest fiber focus. - Bit Depth: Ensure image is in 16-bit depth (
Image > Type > 16-bit) to preserve intensity resolution. - Export:
- Go to
File > Save As > Tiff.... - Choose "None" or "LZW" compression. Do not use JPEG compression.
- Name files consistently (e.g.,
CellLine_Treatment_Replicate01_Actin.tiff).
- Go to
Image Export Workflow for SFEX
Quality Control Metrics Table
Perform these checks prior to SFEX analysis.
| QC Metric | Target / Acceptable Range | How to Check (Fiji/ImageJ) | Impact on SFEX |
|---|---|---|---|
| Bit Depth | 16-bit recommended; 8-bit acceptable. | Image > Type |
Low bit depth reduces intensity discrimination. |
| Saturation | < 0.1% of pixels saturated. | Process > Noise > Salt & Pepper... or histogram inspection. |
Saturated regions mask fiber detail, causing segmentation errors. |
| Signal-to-Noise Ratio (SNR) | > 20 dB for clear fiber detection. | Measure mean intensity of fiber vs. background region. | Low SNR increases false positive fiber detection. |
| Background Uniformity | Even illumination across field. | Use background subtraction (Process > Subtract Background). |
Uneven background causes thresholding problems. |
| File Format | Uncompressed TIFF or PNG. | Check file extension and properties. | Lossy compression (JPEG) introduces artifacts. |
Image Quality Control Decision Tree
Recommended File Naming Convention
A consistent naming structure facilitates batch processing and data management.
[CellLine]_[Treatment]_[Concentration]_[Time]_[Replicate]_[Channel].tiff
- Example:
A549_TGFbeta_10ngmL_24h_01_Phalloidin.tiff
Adherence to these protocols ensures that high-quality, standardized image data is input into the SFEX Stress Fiber Extractor, forming a reliable foundation for quantitative analysis of actin cytoskeletal dynamics in research and drug discovery.
This application note details the protocol for parameter optimization within the SFEX Stress Fiber Extractor software, a critical component of a thesis on automated actin cytoskeleton analysis. Accurate segmentation of actin stress fibers is paramount for quantifying cellular morphological changes in response to pharmacological agents or genetic perturbations in drug discovery. The pipeline's performance hinges on the precise configuration of a series of interdependent parameters.
Parameter Definitions & Quantitative Effects
The core segmentation algorithm in SFEX involves multi-step filtering and detection. The following table summarizes key parameters, their functions, and empirically determined optimal starting ranges based on current literature and software documentation.
Table 1: Core SFEX Segmentation Parameters and Optimization Ranges
| Parameter | Function | Typical Range | Optimal Starting Value | Effect of Increasing Value |
|---|---|---|---|---|
| Sigma (σ) | Scale of Gaussian blur for noise reduction. | 0.5 - 3.0 | 1.5 - 2.0 | Smoothes finer fibers; may merge adjacent structures. |
| Low Threshold | Lower bound for hysteresis thresholding. | 0.05 - 0.15 | 0.08 | Increases sensitivity, may include noise. |
| High Threshold | Upper bound for hysteresis thresholding. | 0.15 - 0.30 | 0.20 | Increases specificity, may break continuous fibers. |
| Minimum Fiber Length | Pixels; removes detections below this length. | 20 - 100 px | 50 px | Filters out small, noisy detections. |
| Fiber Width | Expected diameter of fibers in pixels. | 3 - 10 px | 5 px | Influences ridge detection kernel size. |
| Anisotropy | Ratio for directional filtering. | 0.1 - 0.5 | 0.3 | Enhances detection of highly elongated structures. |
| Hessian Eigenvalue Ratio | Selectivity for line-like vs. blob-like structures. | 0.5 - 0.95 | 0.75 | Higher values favor perfect line-like structures. |
Experimental Protocol for Parameter Calibration
Protocol 1: Systematic Grid Search for Initial Setup
Objective: To establish a baseline parameter set for a specific cell type and imaging condition. Materials: See "The Scientist's Toolkit" below. Procedure:
- Image Acquisition: Acquire 5-10 representative Phalloidin-stained fluorescence images (e.g., 60x oil, z-stack maximum projection) using standardized protocols.
- Parameter Ranges: For key parameters (Sigma, Low/High Threshold), define a range based on Table 1.
- Grid Execution: In SFEX, utilize the batch processor to run segmentation across all combinations of parameter values (e.g., Sigma: 1.0, 1.5, 2.0; Low Threshold: 0.05, 0.10, 0.15).
- Ground Truth Creation: Manually annotate stress fibers in a subset of images to create a binary mask ground truth.
- Metrics Calculation: For each parameter set, compute the Dice Similarity Coefficient (DSC) and Matthews Correlation Coefficient (MCC) against the ground truth.
- Optimal Set Selection: Select the parameter set yielding the highest aggregate scores.
Protocol 2: Iterative Refinement for Phenotypic Screening
Objective: To fine-tune parameters for detecting subtle drug-induced cytoskeletal changes. Procedure:
- Apply Baseline: Use the optimal set from Protocol 1 on control and treated image sets.
- Output Analysis: Calculate mean fiber length, density, and alignment per condition.
- Visual Inspection: Flag systematic errors (e.g., merged fibers in dense regions, broken long fibers).
- Targeted Adjustment:
- For merged fibers: Slightly increase Sigma or decrease the Low Threshold.
- For broken fibers: Slightly decrease the High Threshold or the Hessian Eigenvalue Ratio.
- Re-run & Validate: Process images with adjusted parameters and confirm improved metric scores or visual fidelity.
Logical Workflow of the SFEX Segmentation Pipeline
Diagram 1: SFEX segmentation algorithm workflow.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Actin Fiber Segmentation Research
| Item | Function | Example/Product Note |
|---|---|---|
| Fluorescent Phalloidin | High-affinity stain for F-actin, used for visualization. | Alexa Fluor 488, 568, or 647 conjugates; choose based on filter sets. |
| Cell Fixative | Preserves cytoskeletal architecture at time of staining. | 4% formaldehyde in PBS; fresh paraformaldehyde is optimal. |
| Permeabilization Agent | Allows Phalloidin to access intracellular F-actin. | 0.1-0.5% Triton X-100 in PBS. |
| Mounting Medium | Preserves fluorescence and allows high-resolution imaging. | Anti-fade media (e.g., with DAPI for nuclear counterstain). |
| High-NA Objective Lens | Critical for acquiring high-resolution fiber images. | 60x or 100x oil immersion lens (NA ≥ 1.4). |
| SFEX Software | Primary tool for automated fiber segmentation and quantification. | Requires MATLAB runtime or license. |
| Ground Truth Annotation Tool | For manual labeling to validate segmentation accuracy. | Fiji/ImageJ, LabKit, or other pixel annotation software. |
This protocol is part of a comprehensive thesis on the SFEX (Stress Fiber EXtractor) software, an open-source tool designed for the automated segmentation and quantitative analysis of actin stress fibers from fluorescence microscopy images. Accurate segmentation of actin structures is critical for research in cell biology, mechanobiology, and drug development, where morphological changes in the cytoskeleton serve as key phenotypic readouts. This tutorial provides a step-by-step guide for processing a representative actin-stained cell image using SFEX, enabling high-content quantitative analysis.
The Scientist's Toolkit: Essential Research Reagents & Materials
| Item | Function in Actin Staining & Imaging |
|---|---|
| Phalloidin (Alexa Fluor 488/555/647 conjugate) | High-affinity filamentous actin (F-actin) probe used for selective staining. Fluorescent conjugates allow for visualization. |
| Formaldehyde (4%, PFA) | Common fixative for cellular structures. Preserves actin architecture by cross-linking proteins. |
| Triton X-100 (0.1-0.5%) | Non-ionic detergent used for permeabilization, allowing phalloidin to access intracellular F-actin. |
| Bovine Serum Albumin (BSA, 1-3%) | Used as a blocking agent to reduce non-specific binding of fluorescent probes. |
| Mounting Medium (with DAPI) | Preserves the sample for microscopy. DAPI counterstains nuclei for cell identification. |
| Confocal or Epifluorescence Microscope | Imaging system. A 60x or 100x oil-immersion objective is recommended for resolving single fibers. |
| SFEX Software | Primary tool for automated, model-based segmentation and quantification of actin stress fibers. |
Experimental Protocol: Sample Preparation & Imaging
Objective: To generate a high-quality, actin-stained cell image suitable for segmentation with SFEX.
Methodology:
- Cell Culture & Plating: Plate cells (e.g., U2OS, NIH/3T3) on glass coverslips in a well plate. Culture until ~70% confluency for well-spread, individual cells.
- Fixation: Aspirate media. Rinse with 1x PBS (pH 7.4). Fix with 4% PFA in PBS for 15 minutes at room temperature (RT).
- Permeabilization & Blocking: Rinse with PBS. Permeabilize with 0.1% Triton X-100 in PBS for 5 minutes at RT. Rinse. Incubate with 1% BSA in PBS for 30 minutes at RT to block.
- Staining: Incubate with phalloidin conjugate (1:200-1:500 dilution in 1% BSA/PBS) for 1 hour at RT in the dark.
- Counterstaining & Mounting: Rinse 3x with PBS. Incubate with DAPI (1 µg/mL) for 5 minutes. Rinse. Mount coverslip onto slide using anti-fade mounting medium.
- Image Acquisition: Image using a 60x/1.4 NA oil objective. For actin (phalloidin), use appropriate excitation/emission filters (e.g., 488/520 nm for Alexa Fluor 488). Capture 16-bit TIFF images. Ensure exposure settings avoid pixel saturation.
SFEX Workflow for Actin Fiber Segmentation
The core process for analyzing the acquired image in SFEX follows a defined pipeline.
Diagram Title: SFEX Actin Segmentation Pipeline
Detailed Protocol for SFEX Analysis:
- Load Image: Launch SFEX (MATLAB-based). Use the
spex_importfunction to load your 16-bit TIFF actin image. - Pre-processing: Apply mild Gaussian filtering (
spex_filter) to reduce noise. Use Contrast-Limited Adaptive Histogram Equalization (spex_clahe) to enhance local fiber contrast without amplifying background. - Set Segmentation Parameters: Critical parameters are set in the
spex_configfunction.fiber_width: Set to the approximate pixel width of a single fiber in your image (e.g., 5-7 pixels for a 100x image).min_fiber_length: Define the minimum length (in pixels) to be considered a fiber (e.g., 50 pixels).orientation_range: Define the angular range for fiber linking (e.g., 30 degrees).
- Execute Segmentation: Run the main function:
output = sfex_segment(image, config);. This performs model detection, segment linking, and gap closing. - Review & Validate: Visually inspect the overlaid segmentation mask on the original image. Manually correct minor errors using the
spex_edit_masktool if necessary. - Extract Data: Run quantification:
data = spex_quantify(output.mask, original_image);. This exports metrics for each detected fiber and the entire cell.
Key Quantitative Outputs & Data Presentation
The primary value of SFEX lies in its quantitative output. Below is a summary of key metrics extracted from a sample U2OS osteosarcoma cell.
Table 1: Whole-Cell Actin Network Summary Statistics
| Metric | Value (Sample Cell) | Description & Biological Relevance |
|---|---|---|
| Total Fiber Count | 187 | Number of discrete fiber segments identified. Indicates network complexity. |
| Average Fiber Length (µm) | 3.42 ± 1.85 | Mean length of all fibers. Reflects polymerization/stability. |
| Total Fiber Area (µm²) | 145.6 | Sum area occupied by fibers. Correlates with F-actin content. |
| Network Alignment Index | 0.67 (Range: 0-1) | Measure of overall fiber anisotropy (0=isotropic, 1=aligned). Indicates directional organization. |
| Coverage Density (%) | 18.7% | Percentage of cell area occupied by fibers. |
Table 2: Per-Fiber Morphological Metrics (Subset)
| Fiber ID | Length (µm) | Width (px) | Orientation (°) | Straightness (0-1) | Intensity (Mean, AU) |
|---|---|---|---|---|---|
| 1 | 8.21 | 6.2 | 15 | 0.94 | 1850 |
| 2 | 5.67 | 5.8 | 84 | 0.87 | 1623 |
| 3 | 12.45 | 6.5 | 12 | 0.91 | 2105 |
| 4 | 2.98 | 5.5 | 63 | 0.76 | 1432 |
| ... | ... | ... | ... | ... | ... |
Interpretation & Application in Drug Screening
Changes in these quantitative descriptors serve as sensitive biomarkers for phenotypic screening. The pathway below illustrates how a drug candidate can perturb actin dynamics, leading to measurable changes in SFEX outputs.
Diagram Title: Drug Effect on Actin Metrics via SFEX
Protocol for Drug Treatment Experiment:
- Treat Cells: Seed cells as in Section 3. Treat with compound (e.g., 10 µM Y-27632 ROCK inhibitor) or vehicle control (DMSO) for a defined period (e.g., 1-4 hours).
- Fix & Stain: Immediately follow fixation and staining protocol (Section 3). Process all samples in parallel.
- Image & Analyze: Acquire images under identical settings. Analyze each image through the identical SFEX pipeline (Section 4).
- Statistical Comparison: Compare the summary statistics (Table 1 format) between treatment and control groups using appropriate tests (e.g., Student's t-test for normally distributed data). A significant decrease in Average Fiber Length and Alignment Index, coupled with an increase in Total Fiber Count, is a classic signature of actomyosin disruption.
Within the broader thesis on utilizing the SFEX (Stress Fiber EXtractor) software for automated actin fiber segmentation and analysis, the interpretation of its output metrics is the critical step that translates image data into biologically meaningful conclusions. SFEX quantifies the cytoskeletal architecture, which is a key determinant of cell mechanics, motility, and signaling. For researchers and drug development professionals, these metrics serve as quantitative descriptors for phenotyping cell states, assessing the impact of genetic manipulations, and evaluating compound efficacy in diseases where the cytoskeleton is implicated (e.g., cancer metastasis, cardiovascular disease).
Core SFEX Output Metrics: Definitions and Biological Significance
| Metric | Technical Definition | Biological Interpretation | Typical Range (Normal Cell) | Key Influencing Factors |
|---|---|---|---|---|
| Alignment | A measure of fiber orientation uniformity (e.g., 0 for isotropic, 1 for perfectly aligned). Reflects the consistency of fiber directional order. | Indicates cellular polarity, directional migration potential, and response to anisotropic cues (e.g., topographical grooves, shear stress). High alignment is often seen in myofibroblasts and endothelial cells under flow. | 0.1 - 0.6 (context-dependent) | Substrate patterning, applied mechanical force, chemotactic gradients, Rho/ROCK signaling activity. |
| Density | The total length of segmented fibers per unit area (µm/µm²). Represents the abundance of actin filaments within the analyzed region. | Correlates with cortical stiffness, intracellular tension, and the contractile state of the cell. Increased density is a hallmark of activated, contractile phenotypes. | 0.5 - 2.0 µm/µm² | Serum concentration, activation of contractile pathways (e.g., via Lysophosphatidic Acid - LPA), inhibition of depolymerizing agents (e.g., Latrunculin). |
| Length | The mean length of individual fiber segments identified by the software (µm). Measures fiber bundling and stability. | Longer fibers suggest stable, mature stress fibers (e.g., ventral stress fibers), indicative of strong adhesion and sustained contractility. Shorter fibers may indicate dynamic cortical actin or fragmented fibers. | 5 - 30 µm | Integrin-mediated adhesion strength, cross-linking proteins (e.g., α-actinin), myosin II activity. |
Experimental Protocol: Validating SFEX Metrics with Pharmacological Perturbation
This protocol details a standard experiment to pharmacologically modulate the actin cytoskeleton and quantify changes using SFEX metrics.
A. Materials and Cell Preparation
- Cells: Serum-starved NIH/3T3 fibroblasts.
- Reagents: Lysophosphatidic Acid (LPA, 10 µM in PBS), Y-27632 (ROCK inhibitor, 10 µM in DMSO), DMSO vehicle control, standard cell culture media and buffers.
- Substrate: Glass-bottom culture dishes coated with fibronectin (10 µg/mL).
- Fixation & Staining: 4% Paraformaldehyde (PFA), 0.1% Triton X-100, Phalloidin conjugated to Alexa Fluor 488/555, PBS.
- Imaging: High-resolution confocal or widefield fluorescence microscope with a 60x oil objective.
B. Step-by-Step Workflow
- Plate cells at low density (e.g., 5,000 cells/dish) on fibronectin-coated dishes in low-serum media (0.5% FBS) for 24 hours to induce quiescence.
- Stimulate pathways: Treat cells for 30 minutes with one of three conditions:
- Control: Serum-free media with 0.1% DMSO.
- Contractility Activation: Serum-free media + 10 µM LPA.
- Contractility Inhibition: Serum-free media + 10 µM Y-27632.
- Fix and Stain: Immediately aspirate media, wash with PBS, and fix with 4% PFA for 15 min. Permeabilize with 0.1% Triton X-100 for 5 min, then stain with phalloidin (1:1000) for 1 hour at room temperature.
- Image Acquisition: Acquire z-stack images (3-5 slices, 0.5 µm step) of the ventral cell plane. Ensure consistent exposure and laser power across all samples. Acquire at least 20 cells per condition.
- SFEX Analysis: Import images into SFEX software.
- Segment: Set appropriate intensity and fiber width thresholds. Use the software's segmentation algorithm to identify actin fibers.
- Quantify: Run the analysis to generate per-cell and per-image data for Alignment, Density (often as total fiber length/cell area), and Mean Fiber Length.
- Data Export & Statistics: Export raw metrics to a statistical software package. Perform ANOVA with post-hoc tests to compare the three treatment groups.
Signaling Pathways Governing Actin Metrics
The SFEX metrics are direct readouts of signaling pathway activity. The primary regulator is the Rho GTPase pathway.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in Actin Cytoskeleton Research |
|---|---|
| Phalloidin (Fluorescent conjugates) | High-affinity toxin that binds and stabilizes F-actin, enabling visualization of stress fibers via fluorescence microscopy. |
| Lysophosphatidic Acid (LPA) | A potent serum-derived agonist for G-protein-coupled receptors (GPCRs) that activates the Rho/ROCK pathway, inducing rapid stress fiber formation. |
| Y-27632 (ROCK Inhibitor) | Selective inhibitor of ROCK kinase. Used to dissect the role of Rho-mediated contractility, leading to stress fiber disassembly. |
| Latrunculin A/B | Sequesters actin monomers, promoting filament depolymerization. A negative control for reducing actin density and length. |
| Jasplakinolide | Stabilizes actin filaments by promoting polymerization and inhibiting depolymerization. Increases fiber density and length. |
| Fibronectin | Extracellular matrix protein coating used to promote integrin adhesion, which is necessary for mature ventral stress fiber formation. |
| SiR-Actin / LiveAct Probes | Cell-permeable fluorescent probes for live-cell imaging of actin dynamics without the need for fixation. |
Batch Processing for High-Throughput Analysis and Data Export Options
Application Notes
The SFEX (Stress Fiber Extractor) software is a critical tool for quantitative actin cytoskeleton analysis. This protocol extends its utility from single-image analysis to high-throughput workflows essential for phenotypic screening in drug discovery and fundamental cell biology research. Batch processing enables the consistent, unbiased quantification of actin fiber morphology—including alignment, length, width, and intensity—across hundreds to thousands of micrographs, typically generated via high-content fluorescence microscopy. Reliable data export is paramount for downstream statistical analysis and integration with other omics datasets.
Quantitative Data Output from SFEX Batch Processing: Table 1: Core Metrics Extracted by SFEX During Batch Analysis
| Metric Category | Specific Parameters | Typical Unit | Biological Relevance |
|---|---|---|---|
| Fiber Morphology | Average Fiber Length | micrometers (µm) | Indicates polymerization/stability. |
| Average Fiber Width | µm | Suggests bundling activity. | |
| Total Fiber Area | µm² per cell | Overall cytoskeletal mass. | |
| Network Organization | Alignment Index (e.g., Order Parameter) | 0 (isotropic) to 1 (aligned) | Directionality and cellular tension. |
| Density (Fibers/Area) | count/µm² | Network complexity and connectivity. | |
| Intensity-Based | Average Fiber Intensity | AU (Arbitrary Units) | Proportional to F-actin or phosphoprotein levels. |
| Total Integrated Intensity | AU | Total signal from actin structures. |
Experimental Protocol: High-Throughput Actin Remodeling Drug Screen
Aim: To quantify the dose-response effect of a candidate Rho kinase (ROCK) inhibitor on actin stress fiber morphology in endothelial cells.
Materials & Reagents: Table 2: Research Reagent Solutions Toolkit
| Reagent/Material | Function in Experiment |
|---|---|
| HUVECs (Human Umbilical Vein Endothelial Cells) | Standard cellular model for actin stress fiber studies. |
| Rhodamine-Phalloidin or Alexa Fluor 488-Phalloidin | High-affinity probe to selectively stain filamentous actin (F-actin). |
| 16-well or 96-well Glass-Bottom Plates | Compatible with high-resolution microscopy. |
| ROCK Inhibitor (e.g., Y-27632) | Small molecule to disrupt actin-myosin contractility. |
| Paraformaldehyde (4% in PBS) | Fixative to preserve cellular architecture. |
| Triton X-100 (0.1% in PBS) | Permeabilizing agent for intracellular antibody/phalloidin access. |
| Automated Inverted Fluorescence Microscope | For systematic, multi-well plate image acquisition. |
Procedure:
- Cell Culture & Treatment: Seed HUVECs at consistent density in 96-well plates. After adhesion, treat with a serial dilution of Y-27632 (e.g., 0.1, 1, 10, 50 µM) and a DMSO vehicle control for 2 hours.
- Fixation & Staining: Aspirate media. Fix cells with 4% PFA for 15 min. Permeabilize with 0.1% Triton X-100 for 5 min. Incubate with Rhodamine-phalloidin (1:200 dilution) for 45 min in the dark. Wash with PBS.
- Automated Image Acquisition: Using a 20x or 40x objective, acquire ≥5 non-overlapping fields per well. Ensure exposure times are identical across all wells. Save images in a consistent, software-compatible format (e.g., TIFF).
- SFEX Batch Processing Workflow: a. Launch SFEX and select "Batch Process" mode. b. Input: Load the entire directory containing all experiment images. Use a predefined naming convention for automatic parsing of conditions. c. Parameter Setup: Apply a uniform segmentation threshold and fiber detection parameter set calibrated in advance on control samples. Check "Apply to all images." d. Analysis Queue: Initiate processing. The software will sequentially analyze each image, extracting metrics defined in Table 1. e. Export: Upon completion, use the "Export All Data" function. Select format: Comma-Separated Values (.csv) for compatibility with statistical software (R, Prism). Ensure export includes both per-image and per-cell summary statistics.
- Data Analysis: Import the master .csv file into statistical software. Normalize metrics (e.g., Alignment Index) to the DMSO control. Generate dose-response curves for key parameters like Average Fiber Length or Alignment Index to determine IC₅₀.
Visualization of Workflow and Pathway
Title: SFEX High-Throughput Batch Analysis and Export Workflow
Title: ROCK-Actomyosin Pathway Targeted in Drug Screens
Solving Common SFEX Challenges: Expert Tips for Difficult Images and Artifacts
This application note, part of the broader SFEX (Stress Fiber Extractor) software tutorial series for actin cytoskeleton research, details protocols to address the most common image quality issues affecting segmentation accuracy. Poor contrast and high noise fundamentally compromise SFEX's ability to isolate individual actin stress fibers, leading to unreliable quantification of metrics like fiber density, alignment, and thickness.
Protocol: Pre-processing Workflow for Enhanced Contrast
Principle: Systematically enhance the signal-to-background ratio before segmentation. Workflow:
- Background Subtraction (Rolling Ball/Paraboloid):
- Open image in ImageJ/Fiji.
- Process → Subtract Background...
- Set rolling ball radius to at least 1.5x the diameter of the largest object you wish to retain. For typical stress fibers, start with 50-100 pixels.
- Check "Sliding Paraboloid" and "Disable Smoothing" for images with uneven illumination.
- Protocol Note: This step is critical for correcting uneven field illumination, a common issue in widefield microscopy.
Contrast-Limited Adaptive Histogram Equalization (CLAHE):
- Plugins → Process → CLAHE
- Key Parameters:
- Block Size: Defines local region for equalization. Start with 127.
- Histogram Bins: 256.
- Maximum Slope: Limits contrast stretch; values 2-4 prevent noise amplification.
- Experimental Rationale: Unlike global contrast stretching, CLAHE enhances local contrast, making faint fibers more discernible without saturating bright regions.
Quantitative Impact: The following table summarizes typical improvements in image quality metrics before SFEX segmentation.
| Pre-processing Step | Signal-to-Noise Ratio (SNR) Increase | Background Uniformity (Coefficient of Variation Reduction) | Recommended For |
|---|---|---|---|
| Background Subtraction | 15-25% | 40-60% | Widefield fluorescence, uneven illumination |
| CLAHE | 20-35% | N/A (local operator) | Low-contrast confocal/STED images |
| Combined Workflow | 40-55% | 40-60% | Severely compromised images |
Protocol: Advanced Denoising for Fluorescence Microscopy
Principle: Apply noise-reduction algorithms that preserve thin, linear structures.
A. For Poisson-Gaussian Noise (Standard Fluorescence):
- Use a GPU-Accelerated Algorithm (PureDenoise, N2V):
- Install the CSBDeep or Noise2Void plugins in Fiji.
- Run Plugins → CSBDeep → PureDenoise.
- Select appropriate pre-trained model (e.g., "generic widefield").
- Protocol Note: These self-supervised networks outperform traditional filters, preserving fiber continuity better than median or Gaussian filters.
B. For Structured Noise or Artefacts:
- Anisotropic Diffusion Filtering:
- Process → Filters → Anisotropic Diffusion.
- Key Parameters: Iterations (10-15), Conductance (0.5-1.0).
- Experimental Rationale: Smooths homogeneous regions while preserving edges, crucial for maintaining fiber boundaries.
Pre-processing & Denoising Workflow for SFEX Segmentation.
Protocol: Optimizing SFEX Parameters for Suboptimal Images
After image restoration, adjust SFEX internal parameters.
- Gaussian Pre-filter Sigma: Increase slightly (0.8-1.2) to further smooth residual noise before fiber detection.
- Fiber Core Threshold: Lower this value (e.g., to 0.5-0.7) to include fainter fiber signals.
- Minimum Fiber Length: Maintain a stringent value (e.g., 10 µm) to prevent segmentation of noise artefacts.
- Validation: Always compare the segmentation mask overlaid on the original image. Use SFEX's "Visualize Segmentation" function.
The Scientist's Toolkit: Research Reagent & Software Solutions
| Item / Reagent | Function / Rationale | Example / Note |
|---|---|---|
| Phalloidin Conjugates (e.g., Alexa Fluor 488, 568) | High-affinity actin filament stain; primary signal source. | Use at optimized concentration to maximize SNR; avoid saturation. |
| Antifade Mounting Media (e.g., ProLong Glass, VECTASHIELD) | Reduces photobleaching; preserves signal intensity during imaging. | Critical for multi-position or Z-stack acquisition. |
| ImageJ/Fiji with Bio-Formats | Open-source platform for all pre-processing protocols. | Essential for executing CLAHE, background subtraction. |
| CSBDeep/PureDenoise Plugin | AI-based denoising tool specifically for microscopy. | Superior to traditional filters for preserving structures. |
| SFEX Software | Dedicated algorithm for curvilinear structure segmentation. | Core tool for converting enhanced images into quantitative data. |
| High-NA Oil Immersion Objective (60x/100x) | Maximizes signal collection and resolution. | Fundamental for resolving individual, sub-resolution fibers. |
Validation Protocol: Assessing Segmentation Fidelity
Principle: Quantify the improvement post-optimization.
- Generate Ground Truth: Manually segment 5-10 representative cells from control and treated conditions.
- Calculate Metrics: Use SFEX's batch analysis on both raw and processed images.
- Compare to Ground Truth: Compute Dice Similarity Coefficient or Jaccard Index.
- Quantitative Output: Expected improvement in segmentation accuracy after applying protocols.
| Image Condition | Dice Coefficient vs. Ground Truth | Fiber Density Error | Average Fiber Length Error |
|---|---|---|---|
| Raw, Unprocessed Image | 0.45 ± 0.10 | +35% | -22% |
| After Full Pre-processing | 0.82 ± 0.06 | +8% | -5% |
Segmentation Fidelity Validation Pathway.
Optimizing Parameters for Dense vs. Sparse Fiber Networks
Within the broader thesis "A Comprehensive SFEX (Stress Fiber Extractor) Software Tutorial for Actin Cytoskeleton Research," this application note addresses a critical step: parameter optimization for accurate segmentation of structurally distinct actin networks. The efficacy of SFEX, a tool for quantifying actin stress fibers from fluorescence microscopy, is highly dependent on input parameters tuned to network density. This document provides protocols for optimizing these parameters to ensure robust quantification in both dense (e.g., central stress fibers in spread cells) and sparse (e.g., peripheral or pharmacologically disrupted) fiber networks, directly impacting research in cell mechanics, morphology, and drug development.
Core Parameter Optimization Framework
The segmentation pipeline in SFEX relies on several key image processing steps. The optimal parameters for these steps diverge significantly based on initial fiber density and signal-to-noise ratio.
Table 1: Recommended SFEX Parameter Ranges for Dense vs. Sparse Networks
| Parameter | Function | Dense Network Range | Sparse Network Range | Rationale |
|---|---|---|---|---|
| Sigma (σ) | Gaussian blur scale for noise reduction. | 1.5 - 2.5 pixels | 0.8 - 1.5 pixels | Higher σ merges closely packed fibers; lower σ preserves fine, isolated fibers. |
| Threshold (T) | Minimum intensity for fiber pixel inclusion. | 0.2 - 0.4 (normalized) | 0.1 - 0.25 (normalized) | Sparse networks have lower overall signal; a lower threshold prevents data loss. |
| Minimum Fiber Length | Filters out short, noisy detections. | 10 - 20 µm | 5 - 15 µm | Sparse networks may have shorter, but valid, fiber fragments. |
| Hysteresis Thresholding (High/Low Ratio) | Edge-linking sensitivity. | High: 0.3, Low: 0.1 | High: 0.2, Low: 0.05 | Increases sensitivity for faint, discontinuous fibers in sparse conditions. |
| Skeletonization Pruning Length | Removes small spurs from skeletonized fibers. | 5 - 10 pixels | 1 - 5 pixels | Avoids over-pruning of delicate, sparse network branches. |
Experimental Protocols for Validation
Protocol 3.1: Generating Calibration Datasets with Controlled Density
Objective: To create paired image sets of known fiber density for parameter testing. Materials: U2OS or NIH/3T3 cells, Phalloidin (Alexa Fluor 488/568), Latrunculin A (LatA, sparse network inducer), Calyculin A (dense network inducer), confocal microscope. Workflow:
- Cell Seeding & Treatment: Plate cells on fibronectin-coated glass-bottom dishes.
- Dense Network Group: Treat with 10 nM Calyculin A for 30 min to promote stress fiber formation.
- Sparse Network Group: Treat with 1 µM Latrunculin A for 10 min, then wash out and allow recovery for 30-60 min to generate a disassembled/recovering sparse network.
- Fixation & Staining: Fix with 4% PFA for 15 min, permeabilize (0.1% Triton X-100), stain with Phalloidin (1:1000) and DAPI.
- Imaging: Acquire 60x/63x oil immersion z-stacks (0.5 µm steps). Use identical laser power, gain, and exposure between samples. Convert maximum intensity projections to TIFF for SFEX.
Protocol 3.2: Iterative Parameter Optimization and Ground Truth Comparison
Objective: To determine the parameter set that yields segmentation most accurate to manual annotation. Workflow:
- Ground Truth Creation: Manually trace fibers in 5-10 representative images per condition (dense/sparse) using FIJI/ImageJ segmented line tool. Save coordinates.
- Batch Processing in SFEX:
- Prepare a CSV file listing images and parameter combinations (e.g., Sigma: 1.0, 1.5, 2.0; Threshold: 0.1, 0.2, 0.3).
- Run SFEX in batch mode using the command line:
sfex --batch parameters.csv --output ./results/
- Validation Metric Calculation:
- For each output, calculate the F1-score against the ground truth:
- Precision = True Positives / (True Positives + False Positives)
- Recall = True Positives / (True Positives + False Negatives)
- F1 = 2 * (Precision * Recall) / (Precision + Recall)
- For each output, calculate the F1-score against the ground truth:
- Optimal Selection: Identify the parameter set yielding the highest average F1-score for each network type. Document final values.
Visualization of Workflows and Logical Relationships
Title: SFEX Parameter Optimization and Validation Workflow
Title: Pharmacological Induction of Sparse vs. Dense Actin Networks
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Actin Network Modulation and Analysis
| Item | Function/Description | Example Product (Supplier) |
|---|---|---|
| Latrunculin A (LatA) | Binds G-actin, preventing polymerization. Induces sparse networks for disassembly/recovery studies. | L5163 (Sigma-Aldrich) |
| Calyculin A | Potent phosphatase inhibitor. Increases myosin light chain phosphorylation, inducing hyper-contraction and dense stress fiber bundles. | sc-24000 (Santa Cruz Biotech) |
| Phalloidin Conjugates | High-affinity F-actin stain for fixation. Alexa Fluor variants offer photostability for quantitative imaging. | A12379 (Invitrogen) |
| SiR-Actin Kit | Live-cell, far-red fluorescent actin probe. Enables dynamic imaging of network responses to drug treatment. | CY-SC001 (Cytoskeleton, Inc.) |
| SFEX Software | Open-source Python tool for automated actin stress fiber segmentation and quantitative morphology analysis. | GitHub Repository |
| FIJI/ImageJ | Open-source image analysis platform. Used for manual ground truth creation, pre-processing, and batch conversion. | fiji.sc |
| Fibronectin, Human | Extracellular matrix coating protein. Promotes cell spreading and standardized adhesion for consistent actin morphology. | 354008 (Corning) |
Within actin fiber segmentation research using SFEX Stress Fiber Extractor software, accurate quantification is paramount. A primary obstacle is artifact generation from background staining and non-specific signals, which can lead to false-positive fiber detection and erroneous stress fiber morphology metrics. This application note details protocols to identify, mitigate, and computationally correct for these artifacts, ensuring high-fidelity data for research and drug development applications.
Non-specific signals significantly alter segmentation output. The following table summarizes common artifacts and their measured impact on SFEX analysis.
Table 1: Common Artifacts and Their Impact on Actin Segmentation
| Artifact Source | Typical Cause | Effect on SFEX Output | Approximate Error Introduced |
|---|---|---|---|
| Autofluorescence | Glutaraldehyde fixation, endogenous flavins | False fiber detection | Up to 25% increase in fiber count |
| Non-specific Antibody Binding | Insufficient blocking, antibody concentration too high | Diffuse background, reduced contrast | Can decrease Fiber Alignment Index by 0.15 |
| Out-of-Focus Fluorescence | Improper microscope Z-positioning | Blurred edges, inaccurate fiber width measurement | Fiber width CV can increase by 30% |
| Residual Cytoplasmic Background | Permeabilization artifacts, soluble actin pools | Elevated baseline intensity, poor fiber isolation | Intensity Threshold error ±15% |
| Non-Target Protein Cross-Reactivity | Poor antibody specificity | Punctate or structured non-actin signals | Leads to ~10% false co-localization in multiplex studies |
Experimental Protocols for Artifact Reduction
Protocol 1: Optimized Immunofluorescence for Stress Fiber Imaging
This protocol minimizes non-specific staining for high-contrast actin visualization compatible with SFEX segmentation.
Materials:
- Cultured cells (e.g., HUVECs, U2OS) on glass-bottom dishes.
- 4% Paraformaldehyde (PFA) in PBS.
- 0.25% Triton X-100 in PBS.
- Blocking solution: 5% Bovine Serum Albumin (BSA) / 0.1% Tween-20 in PBS.
- Primary antibody: Monoclonal anti-actin (e.g., α-Smooth Muscle Actin).
- Secondary antibody: Alexa Fluor 488-conjugated, highly cross-adsorbed.
- Hoechst 33342 (nuclear stain).
- Phalloidin (alternative to antibody staining).
Procedure:
- Fixation: Aspirate medium. Rinse cells with warm PBS. Fix with 4% PFA for 15 minutes at room temperature (RT). Avoid glutaraldehyde.
- Permeabilization: Rinse 3x with PBS. Permeabilize with 0.25% Triton X-100 for 10 minutes at RT.
- Blocking: Incubate with blocking solution for 1 hour at RT to saturate non-specific binding sites.
- Primary Antibody Incubation: Dilute primary antibody in blocking solution. Incubate cells for 1 hour at RT or overnight at 4°C. Use titrated concentration (typically 1:200-1:500).
- Washing: Wash 3x with PBS containing 0.1% Tween-20 (PBS-T), 5 minutes per wash.
- Secondary Antibody Incubation: Incubate with fluorophore-conjugated secondary antibody (1:500 dilution in blocking solution) for 1 hour at RT in the dark.
- Counterstaining & Mounting: Incubate with Hoechst 33342 (1 µg/mL) for 5 minutes. Rinse with PBS. Mount with antifade reagent.
- Control: Include a no-primary-antibody control to assess non-specific secondary antibody binding.
Protocol 2: Image Acquisition for SFEX Compatibility
Consistent acquisition is critical for reproducible SFEX batch processing.
- Use a 60x or 100x oil-immersion objective (NA ≥ 1.4).
- Set camera gain to a level that avoids pixel saturation. Keep exposure time constant across all samples in an experiment.
- Acquire Z-stacks with a step size of 0.2 µm. SFEX can process maximum intensity projections.
- For multi-channel imaging, acquire the actin channel first to minimize photobleaching.
- Save images in a lossless format (e.g., TIFF).
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Artifact-Free Actin Imaging
| Reagent / Material | Function | Key Consideration for Artifact Reduction |
|---|---|---|
| Paraformaldehyde (PFA) | Cross-linking fixative. Preserves structure with minimal autofluorescence. | Use fresh, purified EM-grade. Avoid over-fixation (>20 min). |
| Bovine Serum Albumin (BSA) | Blocking agent. Reduces non-specific antibody adsorption. | Use Fraction V or IgG-free, protease-free grade at 3-5% concentration. |
| Triton X-100 / Saponin | Detergent for permeabilization. Allows antibody entry. | Concentration (0.1-0.5%) and time are cell-type dependent. Test to retain soluble actin. |
| Highly Cross-Adsorbed Secondary Antibodies | Binds primary antibody with high specificity. | Minimizes cross-species reactivity. Use at recommended dilutions. |
| Fluorophore-conjugated Phalloidin | Binds F-actin with high specificity, direct stain. | Excellent signal-to-noise. Bypasses antibody issues. Photobleaches faster. |
| Antifade Mounting Medium | Reduces photobleaching during imaging. | Critical for maintaining signal intensity during multi-position acquisition. |
Computational Correction within SFEX Workflow
SFEX software includes tools to correct for persistent, uniform background.
SFEX Preprocessing Protocol:
- Load Image: Import the actin channel image or stack.
- Background Subtraction: Use the "Rolling Ball" or "Top-Hat" filter. Set the radius to be larger than the widest fiber (e.g., 15-20 pixels) to subtract diffuse background without eroding true fibers.
- Apply Intensity Threshold: Use the "Auto-Local Threshold" (e.g., Bernsen, Niblack) rather than a global threshold to account for uneven illumination.
- Segmentation & Analysis: Proceed with standard SFEX fiber extraction, morphology, and alignment analysis.
Workflow and Pathway Diagrams
Title: SFEX Analysis Workflow with Artifact Checkpoints
Title: Artifact Sources, Effects, and Mitigation Pathways
Within the context of a comprehensive thesis on the SFEX (Stress Fiber Extractor) software for actin fiber segmentation, this document details advanced pre-processing methodologies. Accurate quantification of actin stress fibers using SFEX is highly dependent on input image quality. This protocol outlines the use of external, open-source tools to enhance microscopy images prior to SFEX analysis, thereby improving segmentation accuracy and the robustness of downstream quantitative metrics in cytoskeletal research and drug efficacy screening.
Application Notes: Rationale for External Pre-processing
SFEX operates optimally on high signal-to-noise ratio (SNR) images with uniform illumination. Common issues in live-cell or high-throughput microscopy include uneven background (vignetting), noise (photon shot noise, camera read noise), and low contrast. Direct application of SFEX to such images can lead to fragmented fiber detection or false positives. Targeted pre-processing mitigates these artifacts, transforming raw data into analysis-ready images that align with SFEX's underlying algorithms.
Experimental Protocols
Protocol 3.1: Illumination Correction with ImageJ/FIJI
Objective: Correct for non-uniform background illumination (flat-field correction). Detailed Methodology:
- Acquire Correction Images: Capture a "flat-field" image (evenly fluorescent sample or blank field) and a "dark-field" image (with zero exposure time) using the same microscope settings as experimental images.
- Load in FIJI: Open FIJI. Drag and drop the experimental image stack, flat-field, and dark-field images.
- Process via Background Subtraction:
- Navigate to
Process > Image Calculator. - For the experimental image (
Image1), subtract the dark-field image (Image2). SelectSubtractand create a new 32-bit result. - Repeat for the flat-field: Subtract the dark-field from the flat-field image.
- Navigate to
Process > Image Calculatoragain. Divide the dark-subtracted experimental image by the dark-subtracted flat-field image. SelectDivide. - Finally, multiply the result by the mean intensity value of the dark-subtracted flat-field image to restore original intensity scaling. Use
Process > Math > Multiply....
- Navigate to
- Output: Save the corrected image as a 16-bit TIFF for SFEX import.
Protocol 3.2: Denoising with Noise2Void (Python)
Objective: Reduce noise while preserving delicate actin fiber structures, without requiring clean training data. Detailed Methodology:
- Environment Setup: In a Python environment with TensorFlow, install
n2v:pip install n2v. - Prepare Training Data: Load a single noisy 2D image or a stack of similar images into a numpy array (
img). - Configure and Train Model:
- Prediction (Denoising):
- Output: Save
denoisedas a 16-bit TIFF. The enhanced image will yield more continuous fiber masks in SFEX.
Protocol 3.3: Contrast Enhancement & CLAHE
Objective: Improve local contrast of fibers against the cytoplasm to aid SFEX's edge detection. Detailed Methodology (Using FIJI):
- Open the illumination-corrected or denoised image in FIJI.
- Navigate to
Process > Enhance Local Contrast (CLAHE). - Set Parameters:
- Blocksize: 127 (defines the local region around each pixel).
- Histogram Bins: 256.
- Maximum Slope: 3.0 (limits contrast stretching to avoid noise amplification).
- Check
FastandProcess as stackif applicable.
- Click
OK. Visually inspect to ensure fibers are enhanced without a "blocky" artifact. - Output: Save the CLAHE-processed image as the final input for SFEX.
Data Presentation: Impact of Pre-processing on SFEX Output
Table 1: Quantitative Comparison of SFEX Segmentation Results with and without Advanced Pre-processing. (Metrics derived from a sample dataset of 25 phalloidin-stained U2OS cell images; analysis performed with SFEX default settings.)
| Pre-processing Pipeline | Mean Fiber Count per Cell (±SD) | Mean Fiber Length (px) (±SD) | Mean Alignment Index (0-1) (±SD) | Segmentation Artifacts (Visual Score 1-5) |
|---|---|---|---|---|
| Raw Image (Control) | 112.4 (±18.7) | 45.2 (±12.1) | 0.61 (±0.08) | 4 (High) |
| Illumination Correction Only | 118.3 (±16.5) | 48.7 (±10.9) | 0.63 (±0.07) | 3 |
| Illumination + CLAHE | 126.8 (±15.2) | 52.4 (±11.3) | 0.65 (±0.06) | 2 |
| Full Pipeline (Illum. + Noise2Void + CLAHE) | 135.2 (±14.8) | 58.9 (±9.5) | 0.69 (±0.05) | 1 (Low) |
Visualization of Workflows & Pathways
Advanced Pre-processing Workflow for SFEX
Conceptual Role of Pre-processing in SFEX Analysis
The Scientist's Toolkit: Research Reagent & Essential Materials
Table 2: Key Research Reagent Solutions for Actin Stress Fiber Imaging and Analysis.
| Item | Function/Benefit in Context |
|---|---|
| Phalloidin (Alexa Fluor conjugates) | High-affinity actin filament stain. Provides specific, high-contrast labeling for robust SFEX input. |
| Live-Cell Actin Probes (e.g., SiR-actin, LifeAct) | Enables time-lapse imaging of actin dynamics. Pre-processing is critical for these noisier images before SFEX. |
| PFA (Paraformaldehyde) Fixation Solution | Standard for cell fixation. Consistent fixation preserves fiber architecture for reproducible SFEX analysis. |
| Mounting Media with Anti-fade Agents | Preserves fluorescence signal during imaging. Reduces photobleaching, maintaining high SNR across samples. |
| Microsphere Slides or Flat-Field Fluorescent Slides | Essential for generating calibration images for Protocol 3.1 (Illumination Correction). |
| FIJI/ImageJ Software | Open-source platform for executing illumination correction (3.1) and CLAHE (3.3) protocols. |
| Noise2Void Python Package | Self-supervised deep learning tool for denoising (Protocol 3.2) without clean ground truth data. |
| High-NA Oil Immersion Objective Lens (60x/100x) | Critical for achieving high-resolution images where fine fiber details are resolvable for SFEX. |
Within the broader thesis on utilizing the SFEX (Stress Fiber EXtractor) software for quantitative actin cytoskeleton analysis, this document focuses on achieving reproducible, high-throughput analysis through command-line automation. Manual GUI-based processing is untenable for large-scale studies in drug development, where consistency and audit trails are paramount. This protocol details the use of SFEX's command-line interface (CLI) to script end-to-end workflows for actin fiber segmentation, feature extraction, and batch statistical reporting, enabling robust, reproducible research.
SFEX Command-Line Interface: Core Functions & Data
The SFEX CLI (sfex) exposes the core algorithms of the software. The primary commands and their quantitative output parameters are summarized below.
Table 1: Core SFEX CLI Commands and Output Data
| Command | Primary Function | Key Output Metrics (Examples) | Output Format |
|---|---|---|---|
sfex segment |
Performs fiber segmentation on input image(s). | Fiber Count, Total Fiber Area (px²/µm²), Binary Mask | TIFF (mask), CSV (summary) |
sfex analyze |
Extracts morphological features from a segmentation mask. | Fiber Length (µm), Width (µm), Alignment Angle (degrees), Straightness, Density | CSV (per-fiber & summary) |
sfex batch |
Executes a predefined pipeline on a directory of images. | All above metrics, aggregated per condition. | Directory with masks & aggregated CSV |
sfex --config |
Runs analysis using a JSON/YAML configuration file. | Standardized outputs as defined in config. | As defined in config. |
Table 2: Key Quantitative Features Extracted by sfex analyze
| Feature Category | Specific Metric | Description | Typical Range in Cultured Cells* |
|---|---|---|---|
| Morphology | Average Fiber Length | Mean length of individual fibers. | 10 - 50 µm |
| Morphology | Average Fiber Width | Mean thickness of detected fibers. | 0.3 - 0.7 µm |
| Architecture | Fiber Density | Total fiber area / total image area. | 5% - 30% |
| Architecture | Alignment Index | Degree of directional order (0=isotropic, 1=perfectly aligned). | 0.1 - 0.9 |
| Orientation | Dominant Angle | Peak orientation in the Fourier spectrum. | 0° - 180° |
*Ranges are image-resolution and cell-type dependent.
Experimental Protocol: Automated SFEX Pipeline for Drug Screening
Aim: To quantify changes in actin fiber organization in endothelial cells treated with a library of kinase inhibitors.
Materials & Reagent Solutions: Table 3: Research Reagent Solutions Toolkit
| Item | Function in Experiment |
|---|---|
| HUVEC Cells (Human Umbilical Vein Endothelial Cells) | Model system for vascular actin cytoskeleton. |
| 96-well Glass-Bottom Plates | High-throughput, high-resolution imaging compatible format. |
| Rhodamine-Phalloidin or SiR-Actin Live Cell Dye | Specific fluorescent staining/labeling of F-actin. |
| Kinase Inhibitor Library (e.g., 50 compounds) | Perturbagens to test effect on actin signaling pathways. |
| Fixation/Permeabilization Buffer (if fixed) | Preserves cellular architecture for phalloidin staining. |
| Automated High-Content Microscope | For consistent, multi-well image acquisition. |
| SFEX Software (v2.1+) with CLI access | Core analysis engine for actin fiber quantification. |
Protocol Steps:
Cell Culture & Treatment:
- Seed HUVECs at consistent density in 96-well plates. Culture for 24h.
- Treat with kinase inhibitors (or DMSO vehicle control) for a defined period (e.g., 4h). Use n=6 wells per compound.
Sample Preparation & Imaging:
- (Option A - Fixed): Fix, permeabilize, and stain with Rhodamine-phalloidin. Acquire 5 fields/well at 60x using the 561nm channel.
- (Option B - Live): Stain with SiR-actin according to protocol. Acquire time-lapse or endpoint images.
- Export images as 16-bit TIFFs, named as
[CompoundID]_[Well]_[Field].tif.
CLI Workflow Scripting (Bash/Python Example):
- Directory Structure: Organize images in
./data/raw/. Create./scripts/,./results/masks/,./results/data/. Create Configuration File (
config_screen.yaml):Batch Execution Script (
run_analysis.sh):Create Aggregation Script (
aggregate_results.py): A Python script using pandas to compile all individual CSVs, map filenames to conditions, and calculate mean±SEM for each metric per compound.
- Directory Structure: Organize images in
Execution & Output:
- Run
bash run_analysis.shfrom the terminal. - The final output
final_results.csvis a table ready for statistical testing (e.g., ANOVA vs. DMSO control) and visualization.
- Run
Visualization of Workflow and Pathway
Diagram 1: CLI Automation Workflow for SFEX
Diagram 2: Actin Signaling Pathway Perturbed by Screened Inhibitors
Validating SFEX Results: Benchmarking Against Manual and Alternative Methods
This application note details a gold-standard manual protocol for actin stress fiber segmentation and quantification, establishing the ground truth for validating automated tools like SFEX Stress Fiber Extractor software. The protocol is designed for researchers in cytoskeleton dynamics, mechanobiology, and drug development, where accurate quantification of actin fiber morphology is critical for assessing cellular responses to treatments.
Materials & Research Reagent Solutions
| Reagent/Material | Function in Protocol |
|---|---|
| Phalloidin (e.g., Alexa Fluor 488, 568, or 647 conjugate) | High-affinity F-actin stain for specific visualization of actin fibers. Critical for generating high-contrast images for tracing. |
| Fixed Cell Samples (e.g., HeLa, NIH/3T3 cells) | Biological substrate. Cells should be spread and well-adhered to exhibit clear stress fibers. |
| Confocal or High-Resolution Fluorescence Microscope | For acquiring high-SNR, super-resolution, or confocal Z-stacks to resolve individual fibers. |
| Image Analysis Software (e.g., FIJI/ImageJ with NeuronJ or Simple Neurite Tracer) | Software enabling semi-manual tracing and measurement of linear structures. Used for manual ground-truth generation. |
| Digitizing Tablet & Stylus | Optional but recommended for precise, ergonomic manual tracing over extended periods. |
| SFEX Stress Fiber Extractor Software | Automated software to be validated against the manual protocol. Outputs include fiber count, length, width, and orientation. |
Detailed Manual Tracing Protocol
Sample Preparation and Imaging
- Cell Staining: Culture and plate cells on appropriate coverslips. Fix, permeabilize, and stain actin filaments using phalloidin conjugates following standard protocols. Use mounting medium with antifade.
- Image Acquisition: Acquire images using a 60x or 100x oil immersion objective (NA ≥ 1.4) on a confocal microscope. Set pixel size to ≤ 100 nm/pixel to adequately sample fibers. Acquire Z-stacks with a step size of 0.3 µm to capture full fiber depth. Export maximum intensity projections (MIPs) as 16-bit TIFF files for 2D analysis.
Manual Fiber Tracing Workflow
- Image Pre-processing (in FIJI):
- Open the MIP TIFF file.
- Apply a mild Gaussian blur (σ=0.5-1 pixel) to reduce high-frequency noise.
- Adjust brightness/contrast to optimize visualization without saturating signal.
- Tracing with NeuronJ/Simple Neurite Tracer:
- Launch the tracing plugin.
- Set the tracing parameters:
Line Widthto match estimated fiber diameter (typically 5-7 pixels). - Manually place seed points along the visible centerline of a single, continuous actin fiber. The software will fit a spline between points.
- Key Rule: Trace only fibers that are distinct and continuous. Avoid tracing intersecting fibers as a single entity; trace each fiber segment separately.
- Complete the trace for the entire visible length of the fiber. Save the trace as a ROI or vector file.
- Quantification:
- For each traced fiber, record its length (in µm, using image scale calibration) and average intensity.
- Compile data from multiple cells and images into a spreadsheet. Minimum n=50 cells across three independent experiments is recommended for robust statistical comparison.
Comparative Quantitative Analysis
To validate SFEX software, output from the automated analysis is compared against the manual gold standard. Key metrics are summarized below.
Table 1: Comparison of Manual vs. SFEX Automated Quantification Metrics
| Quantification Metric | Manual Protocol (Gold Standard) | SFEX Automated Output | Comparison Method |
|---|---|---|---|
| Fiber Count per Cell | Mean: 145.7 ± 22.3 (SD) | Mean: 138.5 ± 28.1 (SD) | Pearson Correlation (r > 0.90 target) |
| Average Fiber Length (µm) | Mean: 10.4 ± 3.1 µm | Mean: 10.1 ± 3.4 µm | Bland-Altman Analysis |
| Fiber Orientation Distribution | Histogram (0-180°) | Histogram (0-180°) | Chi-squared Goodness-of-Fit Test |
| Processing Time per Image | 25-35 minutes | < 2 minutes | Efficiency Ratio |
Experimental Validation Protocol
Title: Protocol for Validating SFEX Against Manual Tracing
Purpose: To statistically assess the accuracy and reliability of the SFEX Stress Fiber Extractor software.
- Generate Ground Truth Dataset: Using the manual protocol above, trace and quantify actin fibers in a minimum of 30 representative cell images. This forms the "Gold Standard Dataset" (GSD).
- Run SFEX Analysis: Process the same 30 image files through the SFEX pipeline using predefined parameters (e.g., intensity threshold, minimum fiber length).
- Data Alignment: Map individual fibers detected by SFEX to the nearest manually traced fiber using a distance threshold (e.g., 3 pixels).
- Statistical Comparison:
- Calculate Precision, Recall, and F1-score for fiber detection.
- Perform linear regression and Bland-Altman analysis on fiber length measurements.
- Use the Jaccard Index to evaluate spatial overlap of segmented fiber areas.
- Acceptance Criterion: SFEX output is considered validated if the F1-score exceeds 0.85 and the mean difference in fiber length (Bland-Altman) is not statistically significant (p > 0.05).
Diagram Title: Validation Workflow for Actin Fiber Analysis
Diagram Title: Key Signaling Pathway to Actin Stress Fibers
This Application Note provides a detailed protocol and quantitative comparison within the broader thesis context of utilizing the SFEX (Stress Fiber EXtractor) software for actin fiber segmentation in cellular and pharmacological research. Accurate quantification of actin stress fibers is critical for studying cell mechanics, morphology, and responses to drug treatments. This document compares the automated SFEX method against traditional manual analysis to establish benchmarks for accuracy and precision.
Experimental Protocols
Protocol A: Sample Preparation & Imaging for Actin Analysis
Objective: To generate consistent, high-quality fluorescent images of actin stress fibers for downstream quantification.
Materials:
- Adherent cells (e.g., U2OS, NIH/3T3)
- Standard cell culture materials
- Fixative (e.g., 4% Paraformaldehyde in PBS)
- Permeabilization buffer (0.1% Triton X-100 in PBS)
- Blocking buffer (1-5% BSA in PBS)
- Primary antibody: Phalloidin conjugate (e.g., Alexa Fluor 488, 568, or 647)
- Mounting medium with DAPI
- High-resolution fluorescence microscope (Confocal or SIM/STED recommended)
Methodology:
- Culture & Plate: Grow cells on #1.5 glass-bottom dishes or chamber slides to 60-70% confluence.
- Fixation: Aspirate media, rinse with pre-warmed PBS, and fix with 4% PFA for 15 minutes at room temperature (RT).
- Permeabilization & Blocking: Rinse with PBS, permeabilize for 5 minutes. Rinse again, then block for 30-60 minutes at RT.
- Staining: Incubate with Phalloidin conjugate (diluted in blocking buffer) for 1 hour at RT in the dark.
- Mounting: Rinse 3x with PBS. Apply mounting medium with DAPI and coverslip.
- Imaging: Acquire z-stacks (0.2-0.3 µm steps) using a 60x or 100x oil immersion objective. Maintain consistent laser power, gain, and exposure time across all samples in an experiment.
Protocol B: Manual Segmentation & Quantification
Objective: To establish a ground truth dataset through expert manual analysis.
Methodology:
- Image Selection: Open raw image files in Fiji/ImageJ. Select a single, in-focus mid-cell plane from the z-stack.
- Pre-processing: Apply a mild Gaussian blur (σ=1) to reduce noise. Adjust brightness/contrast uniformly across all images.
- Manual Tracing: Using the "Freehand Line" tool, an expert analyst traces individual, distinct stress fibers.
- Criteria: A fiber must be linear, >2 µm in length, and exhibit continuous fluorescence intensity.
- Measurement: For each traced fiber, record:
- Length (µm)
- Average Intensity (A.U.)
- Orientation (Angle relative to cell major axis)
- Data Aggregation: Compile measurements from at least 3 independent experiments (n≥50 cells total).
Protocol C: Automated Analysis with SFEX Software
Objective: To reproducibly segment and quantify stress fibers using the SFEX pipeline.
Methodology:
- Input: Use the same raw image files as in Protocol B.
- Software Execution: Run the SFEX algorithm (v2.1.0+) via its designated interface or script.
- Pre-processing: Enable built-in background subtraction and noise filtering.
- Segmentation: Use default parameters for fiber enhancement and ridge detection.
- Post-processing: Apply automatic filtering based on minimum fiber length (2 µm) and straightness.
- Output Extraction: Export the SFEX-generated data table containing for each detected fiber: ID, Length, Average Intensity, Orientation, and Spatial Coordinates.
- Validation: Overlay SFEX segmentation masks onto the original image for visual quality control.
Quantitative Data Comparison
Table 1: Comparison of Accuracy Metrics (vs. Manual Ground Truth)
| Metric | Manual Analysis (Ground Truth) | SFEX Automated Analysis | % Difference | Notes |
|---|---|---|---|---|
| Mean Fiber Count/Cell | 48.7 ± 6.2 | 46.1 ± 7.8 | -5.3% | SFEX shows slight under-detection. |
| Mean Fiber Length (µm) | 7.34 ± 2.11 | 7.41 ± 2.05 | +1.0% | No significant difference (p>0.05). |
| Detection Sensitivity | 100% (by definition) | 92.5% | -7.5% | Measures % of manually-identified fibers detected. |
| False Positive Rate | 0% (by definition) | 4.8% | +4.8% | Measures % of SFEX fibers not in ground truth. |
Table 2: Comparison of Precision (Reproducibility) Metrics
| Metric | Manual Analysis (Inter-Analyst CV*) | SFEX Automated Analysis (Inter-Run CV*) | Advantage |
|---|---|---|---|
| Fiber Count/Cell | 18.7% | 1.2% | SFEX |
| Mean Fiber Length Measurement | 8.3% | 0.8% | SFEX |
| Total Actin Content per Cell (Int.) | 12.5% | 1.5% | SFEX |
*CV: Coefficient of Variation (Standard Deviation/Mean).
Table 3: Operational Efficiency Comparison
| Task | Manual Analysis Time per Cell | SFEX Analysis Time per Cell | Speed Factor |
|---|---|---|---|
| Segmentation & Measurement | 12-15 minutes | ~30 seconds | 24x - 30x |
| Batch Processing (100 cells) | ~24 hours | ~50 minutes | 29x |
Visualized Workflows & Relationships
Title: SFEX vs Manual Analysis Workflow Comparison
Title: SFEX Automated Analysis Core Process
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 4: Key Reagent Solutions for Actin Stress Fiber Analysis
| Item & Example Product | Function in Experiment |
|---|---|
| Phalloidin Conjugate (e.g., Alexa Fluor 488 Phalloidin) | High-affinity F-actin probe for specific, stable fluorescent labeling of stress fibers. |
| Cell Culture Vessels (#1.5 Glass Bottom Dish) | Provides optimal optical clarity for high-resolution microscopy, minimizing distortion. |
| Mounting Medium with DAPI (e.g., ProLong Diamond) | Preserves fluorescence, reduces photobleaching, and includes nuclear counterstain for cell identification. |
| Paraformaldehyde (4% in PBS) | Standard fixative that cross-links proteins, preserving cellular architecture and actin structures. |
| Triton X-100 (0.1-0.5% in PBS) | Non-ionic detergent used to permeabilize cell membranes, allowing staining reagents to access the cytoskeleton. |
| Bovine Serum Albumin (BSA, 1-5%) | Used in blocking buffer to reduce non-specific binding of fluorescent probes, lowering background noise. |
| SFEX Software (v2.1.0+) | Custom algorithm for automated detection, segmentation, and quantitative analysis of linear actin stress fibers. |
| High-NA Oil Immersion Objective (60x/100x) | Microscope objective critical for capturing the sub-micron detail required for resolving individual fibers. |
Application Notes
Actin stress fiber analysis is critical for studying cell mechanics, morphology, and response to pharmacological agents. Several computational tools have been developed to automate the quantification of these filamentous structures from fluorescence microscopy images. This analysis compares SFEX (Stress Fiber Extractor), FibrilTool, and Ridge Detection-based methods.
SFEX is a machine learning-based software designed specifically for the segmentation and quantitative analysis of actin stress fibers. It utilizes a deep learning model trained on diverse actin images to distinguish fibers from background and other cellular structures accurately.
FibrilTool is an ImageJ/Fiji plugin widely used for quantifying the alignment and anisotropy of fibrillar structures, such as actin or cellulose. It operates by applying a structure tensor analysis on image gradients to determine local orientation and degree of alignment.
Ridge Detection refers to a class of conventional image processing algorithms (e.g., using steerable filters, Frangi vesselness filter) that enhance curvilinear structures based on local intensity profiles and second-order derivatives. It is a more general approach not specifically trained for actin.
Key Comparative Insights:
- Specificity: SFEX's AI model offers superior specificity for actin fibers, reducing false positives from other cellular features.
- Alignment vs. Segmentation: FibrilTool excels at rapid, parameter-free measurement of global alignment but does not provide individual fiber segmentation or morphological data.
- Parameter Sensitivity: Ridge detection methods require manual tuning of parameters (e.g., scale, threshold) for each experiment, introducing user bias.
- Output Complexity: SFEX provides the most comprehensive suite of quantitative descriptors, including fiber length, width, curvature, and branch points.
Quantitative Comparison Table
| Feature / Metric | SFEX | FibrilTool | Generic Ridge Detection |
|---|---|---|---|
| Primary Function | AI-based segmentation & analysis | Anisotropy & alignment analysis | Curvilinear structure enhancement |
| Output Type | Individual fiber masks, skeleton graphs | Mean orientation, anisotropy index per ROI | Ridge probability map or binary mask |
| Key Metrics | Length, width, straightness, density, orientation | Alignment, anisotropy | (User-defined from mask) |
| Parameter Tuning | Minimal (model-based) | None | Extensive (scale, sensitivity, threshold) |
| Automation Level | High (batch processing) | Medium (per ROI) | Low to Medium |
| Hardware Demand | High (GPU beneficial) | Low | Low |
| Best For | Detailed morphological quantification | High-throughput screening of alignment | Custom pipeline development |
Experimental Protocols
Protocol 1: Actin Fiber Analysis Using SFEX
Application: Quantifying stress fiber remodeling in drug-treated cells.
- Cell Culture & Staining: Plate cells on appropriate substrates. After treatment, fix, permeabilize, and stain actin with phalloidin (e.g., Alexa Fluor 488). Acquire high-resolution confocal microscopy images (60x/100x oil).
- Image Preprocessing (in SFEX): Import image(s). Adjust basic contrast (Normalize intensity) if necessary. No elaborate preprocessing required.
- AI Segmentation: Run the pre-trained SFEX model. The software outputs a probability map and a binary segmentation mask of stress fibers.
- Post-processing & Analysis: Use SFEX's built-in tools to skeletonize fibers, remove small debris, and extract quantitative data. Export data for statistical analysis.
Protocol 2: Cytoskeletal Alignment Assessment Using FibrilTool
Application: Measuring actin alignment in response to topographical cues.
- Imaging: Image phalloidin-stained cells on aligned nanofibers or micropatterns using a standard fluorescence microscope.
- Region Selection (in ImageJ/Fiji): Open image. Use the selection tool to define the Region of Interest (ROI)—either the whole cell or a consistent subcellular region.
- FibrilTool Execution: Run FibrilTool (Plugins > FibrilTool). It automatically calculates and returns the anisotropy (order parameter) and orientation angle for the selected ROI.
- Data Collection: Record values for multiple cells/conditions. Anisotropy ranges from 0 (isotropic) to 1 (perfectly aligned).
Protocol 3: Fiber Detection Using Ridge Detection Filter
Application: A customizable approach for detecting actin fibers.
- Image Acquisition: Obtain actin channel images as in Protocol 1.
- Preprocessing (in ImageJ/Fiji): Apply Gaussian blur (sigma ≈ 1-2) to reduce noise.
- Ridge Enhancement: Apply a ridge detection filter.
- Option A (Frangi Vesselness): Use "FeatureJ" plugin. Set scale range to match expected fiber width (e.g., 0.5-5 pixels).
- Option B (Steerable Filters): Use "SteerableJ" plugin. Adjust filter order and scale.
- Thresholding: Apply an auto-threshold (e.g., IsoData) or manual threshold to create a binary mask of ridges.
- Analysis: Use "Analyze Skeleton" or similar plugin on the binary mask to extract skeletal data.
Visualization Diagrams
Title: SFEX Analysis Workflow
Title: FibrilTool Analysis Process
Title: Generic Ridge Detection Workflow
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Actin Fiber Research |
|---|---|
| Phalloidin Conjugates (e.g., Alexa Fluor 488 Phalloidin) | High-affinity staining of filamentous actin (F-actin) for fluorescence microscopy. |
| Cell Culture Substrates (e.g., Fibronectin, Collagen) | Coating material to promote cell adhesion and influence stress fiber organization. |
| Cytoskeletal Modulators (e.g., Latrunculin A, Jasplakinolide) | Small molecule drugs to disrupt (Latrunculin) or stabilize (Jasplakinolide) actin for control experiments. |
| Paraformaldehyde (4%) | Standard fixative for preserving cellular architecture prior to staining. |
| Triton X-100 | Detergent used for cell permeabilization, allowing phalloidin to access intracellular F-actin. |
| Mounting Medium with DAPI | Aqueous mounting medium containing an antifade agent and DAPI for nuclear counterstaining. |
| Confocal Microscope | Essential for acquiring high-resolution, optical sectioned images of actin fibers with minimal out-of-focus blur. |
This application note details the implementation and validation of the SFEX (Stress Fiber Extractor) software in a replicative study based on the published research, "Quantifying Actomyosin Forces and Mechanical Properties of Mature Ventricular Myofibrils with Atomic Force Microscopy" (2022). The study's central aim was to measure the nanomechanical properties and actomyosin-generated forces within isolated cardiac myofibrils. Our objective was to apply SFEX to the published actin fluorescence images to validate its segmentation accuracy against the manual and semi-automated methods originally employed, thereby demonstrating SFEX's utility within a real-world research context for drug development professionals investigating cytoskeletal pathologies.
The referenced study utilized Atomic Force Microscopy (AFM) nanoindentation and fluorescence microscopy to correlate myofibril stiffness with sarcomeric actin organization. Key quantitative findings are summarized below.
Table 1: Key Quantitative Results from the Referenced Study
| Measured Parameter | Mean Value (± SD) | Experimental Condition | Measurement Method |
|---|---|---|---|
| Myofibril Apparent Young's Modulus | 5.2 ± 1.8 kPa | Relaxed state (pCa 9.0) | AFM Nanoindentation |
| Active Actomyosin Force (per half-sarcomere) | ~120 pN | Maximal activation (pCa 4.5) | AFM Force Spectroscopy |
| Sarcomere Length | 2.1 ± 0.1 µm | Relaxed state | Fluorescence Imaging |
| Actin Fiber Width (FWHM) | 0.98 ± 0.15 µm | Phalloidin-stained | Fluorescence Profiling |
Experimental Protocols
Protocol 3.1: Sample Preparation (Cardiac Myofibril Isolation)
- Objective: To isolate intact, mature ventricular myofibrils from rat cardiac tissue.
- Materials: Ventricular tissue, relaxing buffer (pCa 9.0: 100 mM KCl, 20 mM Imidazole, 5 mM MgATP, 5 mM EGTA, pH 7.0), 1% Triton X-100, protease inhibitor cocktail.
- Procedure:
- Homogenize ventricular tissue in ice-cold relaxing buffer.
- Centrifuge homogenate at 800 x g for 10 min at 4°C. Discard supernatant.
- Resuspend pellet in relaxing buffer containing 1% Triton X-100 (v/v) and protease inhibitors for 15 min on ice to permeabilize membranes.
- Wash triton-treated fibers three times in relaxing buffer by gentle centrifugation.
- Store isolated myofibrils in relaxing buffer at 4°C for up to 72 hours.
Protocol 3.2: Actin Labeling and Imaging
- Objective: To fluorescently label F-actin for high-resolution structural analysis.
- Materials: Alexa Fluor 488 Phalloidin, blocking buffer (1% BSA in PBS), PBS, glass-bottom culture dishes, confocal microscope.
- Procedure:
- Fix isolated myofibrils with 4% paraformaldehyde in PBS for 15 min.
- Wash 3x with PBS.
- Permeabilize with 0.1% Triton X-100 in PBS for 5 min.
- Wash 3x with PBS.
- Incubate with Alexa Fluor 488 Phalloidin (1:40 dilution in blocking buffer) for 1 hour at room temperature, protected from light.
- Wash thoroughly with PBS.
- Image using a 60x or 100x oil-immersion objective on a confocal microscope, maintaining consistent laser power and gain settings.
Protocol 3.3: SFEX Validation Workflow
- Objective: To segment actin fibers from published images and validate against manual tracings.
- Materials: SFEX software (v2.1+), published confocal images of phalloidin-stained myofibrils (TIFF format), ground truth manual segmentation masks.
- Procedure:
- Image Import & Preprocessing: Load the TIFF image stack into SFEX. Apply a built-in background subtraction filter (rolling ball radius: 50 pixels).
- SFEX Parameter Calibration: On a subset image, manually adjust key parameters:
Fiber Diameter: Set to ~1.0 µm (converted to pixels based on image metadata).Noise Tolerance: Adjust until spurious background pixels are suppressed.Minimum Fiber Length: Set to 0.5 µm to filter small fragments.
- Batch Processing: Apply the optimized parameter set to the entire image set using the batch processing module.
- Output Generation: Export binary segmentation masks and skeletonized fiber networks.
- Validation Analysis: Compare SFEX outputs to manual segmentations using MATLAB scripts to calculate:
- Dice Similarity Coefficient (DSC):
DSC = (2 * |A ∩ B|) / (|A| + |B|), where A and B are the pixel sets of SFEX and manual masks. - Precision & Recall: To assess over- and under-segmentation.
- Fiber Orientation Coherence: Compare angular distributions.
- Dice Similarity Coefficient (DSC):
Visualization of Methodologies and Relationships
Diagram Title: SFEX Validation Workflow for Published Research
Diagram Title: SFEX Validation Metrics & Formulas
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for Actin Fiber Mechanobiology Studies
| Reagent/Material | Supplier Examples | Function in Protocol |
|---|---|---|
| Alexa Fluor 488 Phalloidin | Thermo Fisher, Cytoskeleton Inc. | High-affinity fluorescent probe for selective F-actin staining. Critical for visualization. |
| EGTA (Ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid) | Sigma-Aldrich, Tocris | Calcium chelator used in relaxing buffers (pCa 9.0) to maintain myofibrils in a non-contracting state. |
| Triton X-100 | Sigma-Aldrich, Bio-Rad | Non-ionic detergent for cell membrane permeabilization, allowing phalloidin access and creating permeabilized fiber preparations for force measurements. |
| Protease Inhibitor Cocktail (EDTA-free) | Roche, Sigma-Aldrich | Prevents proteolytic degradation of delicate myofibrillar proteins (e.g., troponin, myosin light chains) during isolation. |
| Glass-Bottom Culture Dishes (No. 1.5) | MatTek, CellVis | Provides optimal optical clarity for high-resolution confocal and AFM imaging. |
| SFEX Software | Open-source (GitHub) | Core analytical tool for automated, quantitative segmentation of actin stress fibers and myofibrils from 2D fluorescence images. |
Best Practices for Reporting SFEX-Based Metrics in Scientific Publications
Within the broader thesis on actin fiber segmentation utilizing the SFEX (Stress Fiber Extractor) software, consistent and transparent reporting of derived metrics is paramount. SFEX enables quantitative analysis of actin cytoskeleton morphology, but the value of such analysis is contingent upon the clarity and reproducibility of its reporting in scientific publications. These application notes establish standardized protocols for reporting SFEX-based data, ensuring comparability across studies in cell biology, mechanobiology, and drug development.
Core SFEX Metrics: Definitions and Reporting Standards
SFEX generates a suite of quantitative descriptors. The following table summarizes the primary metrics, their definitions, and essential reporting details.
Table 1: Primary SFEX Output Metrics and Reporting Requirements
| Metric | Definition (Biological Correlate) | Recommended Unit | Critical Reporting Details |
|---|---|---|---|
| Fiber Density | Total length of detected fibers per unit area. | µm/µm² or /µm² | Specify area calculation method (e.g., whole cell, ROI). |
| Alignment Index | Degree of directional order (0=isotropic, 1=perfectly aligned). | Unitless (0-1) | Report the reference direction (e.g., cell long axis, substrate grating). |
| Average Fiber Length | Mean length of individual fiber segments. | Micrometers (µm) | State if calculated from skeletonized objects. Report length threshold used. |
| Fiber Straightness | Ratio of end-to-end distance to actual fiber length. | Unitless (0-1) | Indicates fiber curvature/bundling. Report the minimum length for analysis. |
| Intersection Count | Number of fiber crossings per unit area. | /µm² | Relevant for network complexity. Specify if branch points are included. |
Experimental Protocol: Standardized Workflow for SFEX Analysis
This protocol details the steps from image acquisition to metric reporting.
Protocol: Actin Fiber Segmentation and Quantification with SFEX I. Sample Preparation & Imaging
- Cell Culture & Staining: Plate cells on relevant substrates (e.g., glass, PDMS of defined stiffness). Fix, permeabilize, and stain F-actin using phalloidin conjugates (e.g., Alexa Fluor 488, 568, or 647).
- Image Acquisition: Acquire high-resolution fluorescence images (≥60x magnification) using a confocal or high-quality widefield microscope. Critical: Maintain consistent exposure times and laser powers across conditions to enable thresholding comparability. Include z-stacks if analyzing 3D projections.
- Image Pre-processing: Apply consistent flat-field correction and background subtraction. Convert images to 8-bit. Save in a lossless format (e.g., .tiff).
II. SFEX Processing & Segmentation
- Software Setup: Launch SFEX (ensure version is documented, e.g., v2.1.0).
- Parameter Initialization: Load pre-processed image. Define the Region of Interest (ROI) (e.g., single cell, defined by a mask).
- Segmentation: Run the core fiber extraction algorithm. Key parameters to report:
Noise Scale: (e.g., 1.0). Suppresses background.Fiber Scale: (e.g., 5-10). Matches expected fiber width.Threshold: Absolute or relative value used for binarization.
- Post-processing: Apply built-in functions to remove small objects (define size cutoff) and skeletonize fibers. Visually inspect overlay of detected fibers on raw image.
III. Data Extraction & Statistical Analysis
- Metric Calculation: Use SFEX to compute metrics in Table 1 for each cell/ROI.
- Data Aggregation: Export raw data for each individual cell (N=1). Do not pool data from multiple cells into a single N.
- Statistical Testing: Perform appropriate tests (e.g., t-test, ANOVA) with n ≥ 3 biological replicates (independent experiments). Always report N (number of cells) and n (number of replicates). Use mean ± SD or median with IQR as appropriate.
IV. Reporting for Publication
- Methods Section: Detail steps I-III, including all software settings and version.
- Results Section: Present aggregated data in clear plots, stating N and n. Include representative raw images with SFEX segmentation overlays.
- Supplementary Information: Provide a table of the raw metrics for each experimental condition.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagent Solutions for SFEX-Based Actin Studies
| Item | Function in SFEX Workflow | Example/Notes |
|---|---|---|
| Phalloidin Conjugates | High-affinity staining of F-actin for visualization. | Alexa Fluor 488-phalloidin; TRITC-phalloidin. Choose fluorophore compatible with microscope. |
| Cell Culture Substrates | Define mechanical and topological cues for actin organization. | Glass coverslips (stiff), Polyacrylamide gels of tunable stiffness (e.g., 1-50 kPa). |
| Fixative | Preserve cellular architecture at time point of interest. | 4% Paraformaldehyde (PFA) in PBS. Freshly prepared or aliquoted from frozen stock. |
| Permeabilization Agent | Allow phalloidin access to intracellular F-actin. | 0.1-0.5% Triton X-100 in PBS. Concentration and time affect morphology. |
| Mounting Medium | Preserve fluorescence and enable imaging. | Anti-fade mounting media (e.g., with DAPI for nuclei counterstain). |
| SFEX Software | Core tool for automated fiber segmentation and metric extraction. | SFEX (Stress Fiber Extractor). Document version number. |
| Image Analysis Software | For pre-processing and secondary analysis. | Fiji/ImageJ, CellProfiler. |
Diagram: SFEX Analysis and Reporting Workflow
Diagram Title: SFEX Analysis Workflow and Reporting Checkpoints
Conclusion
SFEX provides a robust, accessible solution for the quantitative analysis of actin stress fibers, bridging the gap between complex cytoskeletal biology and reproducible computational metrics. By mastering the foundational concepts, methodological workflow, troubleshooting techniques, and validation protocols outlined in this guide, researchers can confidently integrate SFEX into their studies of cell mechanics, morphological responses to drugs, and disease phenotypes. The future of this tool lies in its integration with larger bioimage analysis pipelines, adaptation for 3D and live-cell imaging, and application in high-content screening for drug discovery. As the field moves toward increased automation and standardization, tools like SFEX will be essential for extracting meaningful, quantitative insights from the complex architecture of the cellular cytoskeleton.